 |
 |
- Search
| Genomics Inform > Volume 21(2); 2023 > Article |
|
Abstract
Adaptation of infections and hosts has resulted in several metabolic mechanisms adopted by intracellular pathogens to combat the defense responses and the lack of fuel during infection. Human tuberculosis caused by Mycobacterium tuberculosis (MTB) is the worldŌĆÖs first cause of mortality tied to a single disease. This study aims to characterize and anticipate potential antigen characteristics for promising vaccine candidates for the hypothetical protein of MTB through computational strategies. The protein is associated with the catalyzation of dithiol oxidation and/or disulfide reduction because of the proteinŌĆÖs anticipated disulfide oxidoreductase properties. This investigation analyzed the protein's physicochemical characteristics, protein-protein interactions, subcellular locations, anticipated active sites, secondary and tertiary structures, allergenicity, antigenicity, and toxicity properties. The protein has significant active amino acid residues with no allergenicity, elevated antigenicity, and no toxicity.
Pathogenic mycobacteria are significant sources of illness in humans and animals. Despite the accessibility of antibiotics and chemotherapeutic candidates that are efficient against some mycobacteria, the emergence of drug-resistant strains necessitates the development of new active molecules and intervention strategies [1-3]. Within more than 130 years since discovering Mycobacterium tuberculosis (MTB) as the causative microorganism responsible for human tuberculosis (TB) by Robert Koch, numerous scientific advances have been made to help cope with this significant pathogen. However, despite this progress, MTB still holds many unresolved secrets. Much work remains to be done to translate the essential findings from recent research into novel strategies against the pathogen [4-7]. Tuberculosis, one of the ancient recorded human diseases, continues to be one of the leading causes of death, claiming two million lives annually. TB affects bone, the central nervous system, and many other physiological systems. However, it is essentially a pulmonary illness caused by the precipitation of aerosolized MTB onto lung alveolar surfaces. From this point, the causes of the illness are contingent on the immunological reactivity of the host to varying degrees [8-10].
MTB has an irregular, highly recurrent life cycle that encompasses a varied and heterogeneous spectrum of habitats and physiologic states, many of which are unique from other infections. It is not unanticipated that MTB has conceived a specific set of metabolic capacities to facilitate its adaption to and movement across hosts, given its peculiar, if not distinctive, environment [11-13].
Numerous advancements in computational biology have made it possible to construct diverse technologies and strategies for predicting protein structure and the identification of sequence commonalities for active investigation and the analysis of active site residue relationships [14-16]. A bioinformatics examination of the proteins enables one to assess their three-dimensional architectural structure, categorize novel features, investigate specific processes to understand our biological lineage, uncover different clusters, and assign the proteinsŌĆÖ function. The obtained information can also convey reasonable pharmacological strategies and assets in developing promising anti-disease medications [17-19]. The hypothetical protein from MTB consists of disulfide oxidoreductase involved in the catalyzation of dithiol oxidation and/or disulfide reduction of target sites in MTB.
The amino acid sequence of the protein was obtained from the NCBI database in FASTA format. The physicochemical properties were determined using ProtParam (ExPASy) and SMS v.2.0 programs. Afterwards, the subcellular location of the selected protein was determined. This study also anticipated the protein family, superfamily, domain, coil, and folding pattern of the protein. The STRING program was used for protein protein interaction determination. Moreover, secondary structural documentation was performed using the SOPMA, DISOPRED (v. 3.0), and SPIPRED (v. 4.0) programs. The tertiary structure was predicted using the Modeller program with the HHpred database and validated by the PROCHECK, Verify3D, and ProSA-web tools. Furthermore, the CASTp server was used for active site determination of the selected protein present in MTB. Additionally, the antigenicity, allergenicity, and toxicity properties of the protein were determined.
The physicochemical parameters of the protein were evaluated by the ProtParam assessment tool of the ExPASy server program [20] and the SMS v.2.0 program (https://www.bioinformatics.org/sms2/index.html, accessed on September 10, 2022).
The STRING program (v.11.5) [33] was used to determine the protein-protein (pr-pr) interaction.
The tertiary structure of the protein is generated by using the Modeller program [37]. The HHpred tool selected the most suitable template for protein structure anticipation [38-40]. The PROCHECK and Verify3D programs of the SAVES (v.6.0) tool were used for the structural validation of the protein [41]. Additionally, the ProSA-web program was used to calculate the Z-score and validate the modeled 3D structure of the protein [42].
The CASTp program was used to determine the active sites in the protein [43].
The proteinŌĆÖs amino acid sequence was retrieved from the NCBI database in FASTA format. The protein contains 173 amino acids (Table 1).
By examining the properties of each amino acid in the protein, it is possible to comprehend how its physicochemical properties were characterized. The ProtParam program estimated the physicochemical characteristics. The protein comprises 173 amino acids, whereas Ala (n = 30, 17.3%) is the most abundant amino acid in the protein sequence (Table 2, Figure 1). There is no His (H) in the protein sequence. The half-life of a protein is defined as the time required for the radio-labeled focal protein concentration to fall by 50% relative to the quantity at the beginning of the chasing [47]. The estimated half-life for the protein of about 30 h (mammalian reticulocytes, in vitro), >20 h (yeast, in vivo), and >10 h (Escherichia coli, in vivo). The demonstrated isoelectric point (pI), the total number of atoms, and molecular weight as of 5.19, (4.98*), 2,572, and 18,382.98 Dalton (Table 2).
Moreover, the total number of positively (Arg + Lys) and negatively charged residues (Asp + Glu) are 10 and 11 in the protein. The instability index (29.40) demonstrates protein stability, whereas the aliphatic index (86.42) denotes protein balance over a broad temperature scale. The grand average of hydropathicity (GRAVY, 0.334) determines the enhancement of thermostability [48].
The computerized estimation of the subcellular location of bacterial proteins is essential for proteome categorization and for selecting novel therapeutic targets and vaccination candidates. Various subcellular localization predictors have been created in recent years, including both generic localized and feature-based predictors [49-52]. The CELLO (v.2.5) and PSORTb (v3.0) predicted the subcellular location of the protein as extracellular (Table 3).
Moreover, transmembrane helix identification in integral membrane proteins is an essential bioinformatics component. In addition to predicting individual transmembrane helices, the most effective approaches to date strive to predict the complete topology of the protein, including the entire count of transmembrane helices and their direction related to the membrane [27,53]. The TMHMM (v.2.0) and HMMTOP (v.2.0) programs anticipated the protein has a single transmembrane helix (at 12ŌĆō30 region in the protein residue). Most membrane proteinsŌĆÖ transmembrane portions have helices as their secondary structures. When a membrane protein is conducting its task, whether sending messages throughout the membrane or assisting an ion channel in unlocking or closing, the transmembrane helices frequently move together [54-57].
The Conserved Domain Database (CDD) intends to annotate biomolecular sequences with the evolutionarily conserved protein domain placement. A repository of pre-computed domain identification is kept for NCBIŌĆÖs Entrez database-tracked proteins, and real-time search facilities are provided. CDD also facilitates comparative analysis of protein families employing conserved domain architectures, and a new curation effort focuses on giving functional categorization of various subfamily structures [58-60]. The CDD tool classified the protein as protein disulfide oxidoreductase (domain architecture ID 10122406, accession ID cd03011) associated with the catalyzation of dithiol oxidation or disulfide reduction of target proteins [61].
The GenomeNet program identified six different motifs, including AhpC-TSA (position between 43-144, independent E-value 6.8 ├Ś 10-15), Redoxin (position between 44ŌĆō137, independent E-value 1.60 ├Ś 10-10), thioredoxin (position between 52ŌĆō114, independent E-value 1.0 ├Ś 10-4), thioredoxin-2 (position between 59ŌĆō164, independent E-value 4.30 ├Ś 10-5), thioredoxin-8 (position between 61-146, independent E-value 3.6 ├Ś 10-4), and thioredoxin-9 (position between 57-108, independent E-value 8.3 ├Ś 10-2). The Pfam program [62] and the ScanProsite tool also validated the six motifs, including AhpC-TSA (accession ID PF00578), Redoxin (accession ID PF08534), thioredoxin (accession ID PF00085), thioredoxin-2 (accession ID PF13098), thioredoxin-8 (accession ID PF13905), and thioredoxin-9 (accession ID PF14595). Moreover, the SuperFamily program anticipated the protein as a member of the thioredoxin-like superfamily (accession ID 52833, E-value 2.1 ├Ś 10-30). Thioredoxins are small proteins composed of around one hundred amino acid residues that participate in numerous redox processes. Thioredoxins operate by reversibly oxidizing an active center disulfide bond. An intramolecular disulfide bond connects the two cysteine residues in reduced or oxidized forms [63-68].
The cellular machinery is supported by proteins as well as their functional connections. For a comprehensive comprehension of biological events, their connection network must be considered, yet the existing knowledge on protein-protein relationships is inadequate and of different annotation granularity and trustworthiness. The STRING database attempts to gather, assess, and integrate all publicly accessible sources of knowledge on protein-protein interactions and supplement them with computational predictions. It seeks to establish a worldwide network that is complete and objective, incorporating both primary (physical) and secondary (functional) linkages [69-72]. The STRING program (v.11.5) was performed to determine the protein-protein (pr-pr) interaction (Fig. 2). The string program demonstrated the functional fellows with the scores as of trxC (0.795), Rv2876 (0.734), dipZ (0.884), Rv1929c (Rv1929c), mpt63 (0.731), Rv1676 (0.818), Rv2968c (0.798), Rv1929c (0.795), Rv2969c (Rv2969c), and Rv1138c (0.763).
The trxC, Rv2876, dipZ, Rv1929c, mpt63, Rv1676, Rv2968c, Rv1929c, Rv2969c, and Rv1138c are the thioredoxin that participates in multiple redox reactions through the reversible oxidation of its active center dithiol to a disulfide and catalyzes dithiol-disulfide exchange reactions, possible conserved transmembrane protein, cytochrome c-type biogenesis protein, conserved hypothetical protein, immunogenic protein mpt63 (antigen mpt63/mpb63), uncharacterized protein, probable conserved integral membrane protein, conserved hypothetical protein (uncharacterized), possible conserved membrane or exported protein, and possible conserved membrane or exported protein, respectively [73-76]. The mtp53 is a soluble secreted antigen mpt53 precursor, disulfide oxidoreductase, that speeds up the oxidation of diminished, unfolded secreted proteins to make disulfide bonds [77].
Static high-resolution structures have contributed significantly to our protein structure and molecular activity knowledge. As structural biology has progressed, it has become evident that high-resolution structures alone cannot adequately represent the molecular basis for the structure and the action of proteins in solution [78-80]. The secondary structural components, such as helix, sheet, coil, and turn, strongly correlate with proteins' operation, architecture, and interaction [81-84]. The SOPMA program identified the alpha-helix (n = 66, 38.15%), extended strand (n = 39, 22.54%), beta-turn (n = 13, 7.52%), and random coil (n = 55, 31.79%) (Fig. 3).
The SPIPRED (v.4.0) and the DISOPRED (v.3.0) programs were used to determine the secondary structure, sequence plot, and transmembrane topology (Fig. 4).
Homology modeling is a technique for constructing the three-dimensional structures of proteins based on their primary sequence and using existing information from structural matches to other proteins. Sequence/structure compatibility is improved in the homology modeling procedure, a framework is constructed, and side chains are appended [85,86]. The HHpred is an accessible, collaborative web service for protein bioinformatics analysis. Experts and non-experts have access to a vast array of integrated outside-generated, cutting-edge bioinformatics tools [87].
The HHpred is a robust technology for remote homology determination and structure prediction, first constructed as hidden Markov models as well as popularized by the first pairwise comparison study of homologous protein patterns. It permits several repositories, such as PDB, CDD, Pfam, SMART, SCOP, and COG [38]. It accepts a single query array or many lineups as an entry and provides the results via a user-friendly layout similar to PSI-BLAST. Local or worldwide integration and the discovery of secondary systems are among the screening capabilities. HHpred can construct multiple inquiry prototypes, various model alignments with numerous schemes, and three-dimensional representations calculated with the Modeller program from these combinations [39]. The most suitable template (HHpred ID: 1LU4_A, PDB ID: 1LU4) was selected with the probability (99.92%), E-value 1.7 ├Ś 10-22, and target length of 136.
Moreover, the PROCHECK program of the SAVES (v.6.0) tool was used for the Ramachandran plot assessment (Fig. 5). The amino acid sequences in the most favored regions, residues in additional allowed regions, the number of glycine residues (shown as triangles), and the number of proline residues is 9 are 106 (91.4%), 10 (8.6%), 8, and 9, respectively. Likewise, the Verify3D program demonstrated as 100% of the residues averaged a 3D-1D score (Ōēź0.2), whereas at least 80% of the amino acids scored (Ōēź0.2) in the 3D/1D profile to pass [88]. Identifying flaws in theoretical and experimental representations of protein architecture is a fundamental challenge in structural biology. ProSA is a well-known application with a broad user base commonly used to improve and assess empirical protein architectures and structure projection and analysis. Protein structural investigation is often a demanding and laborious process [42]. In addition, the ProSA-web calculated the Z-score as -6.53.
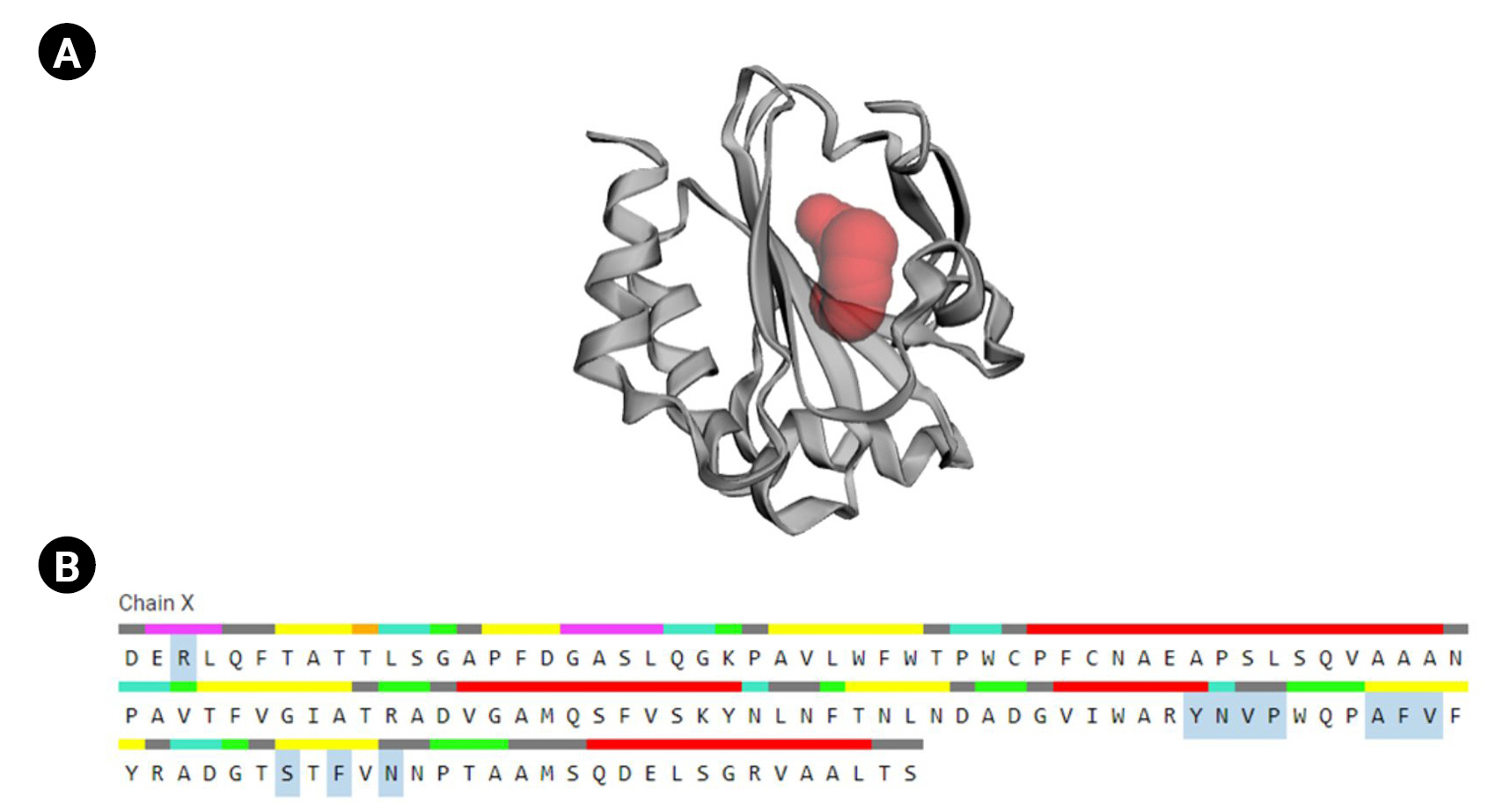
CASTp is a database platform capable of locating zones on proteins, outlining their outlines, determining the dimensions of the regions, and calculating their area. This incorporates pockets on the surface of proteins and hidden vacuums within proteins. The computation includes a pocket and volume spectrum or vacuum, which are mathematically calculated by a solvent-accessible surface (Richards surface) and molecular surface model (surface of Connolly). CASTp could be used to study surface characteristics and protein functioning regions. CASTp delivers a graphical, user-interface-flexible, dynamic display and on-the-fly assessment of user-submitted constructions [43]. The CASTp v.3.0 program demonstrated 11 different active sites in the protein. The highest functioning zones of the modeled protein were recognized between the area of 80.526 and the volume of 37.099 (Fig. 6).
Vaccine development in the post-genomic age often commences with the in silico assessment of genome data, with the most likely defensive antigens anticipated instead of the cultivation of pathogenic bacteria. Despite the apparent benefits of this method, such as speed and cost-effectiveness, its success is contingent on the precision of antigen prediction. Antigens are identified using sequence alignment in most cases [89-93]. This situation is hazardous for several reasons. Specific proteins may share comparable structures and biological activities despite visible sequence similarity. The antigenicity of a sequence may be encoded in a subtle and convoluted manner, making straightforward detection by sequence alignment impossible [94-96]. Considering the proteinŌĆÖs physical and chemical attributes, the VaxiJen program projected that it was antigenic, with the baseline threshold of 0.4 used as the antigenicity parameter. The overall anticipated antigenicity score was measured as 0.5936.
Allergy overreaches the immune function to a formerly exposed, normally innocuous chemical, leading to skin rash, mucous membrane swelling, sneezing or wheezing, or other aberrant symptoms. The rising prevalence of altered proteins in food, commercial items, laundry detergent, medical therapies, and diagnostics renders anticipating and detecting possible allergies a significant social concern. Using bioinformatics, allergen prediction has been extensively studied, and several tools have been created over the past decade; many are accessible on the complimentary internet [97,98]. Furthermore, the AllerTOP (v. 2.0) anticipated the protein as of probable non-allergen protein. Over the last several decades, scientific study has focused on developing peptide/protein-based treatments for various ailments. With various benefits over small molecules, including high selectivity, significant penetration, and simplicity of production, peptides have emerged as prospective therapeutic agents against various disorders. However, the toxicity of peptide- and protein-based therapies is one of their limitations. To forecast the toxicity of peptides and proteins, we built in silico models in this work [99-101]. The ToxinPred program predicted the protein as nontoxic.
Adaptation between pathogens and their innholders has resulted in several metabolic strategies employed by intracellular infections to cope with the defense responses and nutritional insufficiencies throughout infection. Comprehending how proteins act is essential for explaining how they operate, and this protein contains disulfide oxidoreductase, a crucial enzyme associated with the oxidation of dithiol and/or the reduction of disulfide in target sites. This study reveals the fundamental characteristics of the protein of MTB. Moreover, the protein-protein interactions, active amino acid residues, allergenicity, antigenicity, and toxicity uncover the protein potentiality of MTB infection.
Fig.┬Ā1.
Amino acid composition. The protein contains Ala (30, 17.3%), Arg (7, 4.0%), Asn (10, 5.8%), Asp (8, 4.6%), Cys (2, 1.2%), Gln (6, 3.5%), Glu (3, 1.7%), Gly (10, 5.8%), Ile (5, 2.9%), Leu (13, 7.5%), Lys (3, 1.7%), Met (4, 2.3%), Phe (12, 6.9%), Pro (11, 6.4%), Ser (12, 6.9%), Thr (12, 6.9%), Trp (5, 2.9%), Tyr (3, 1.7%), and Val (17, 9.8%).
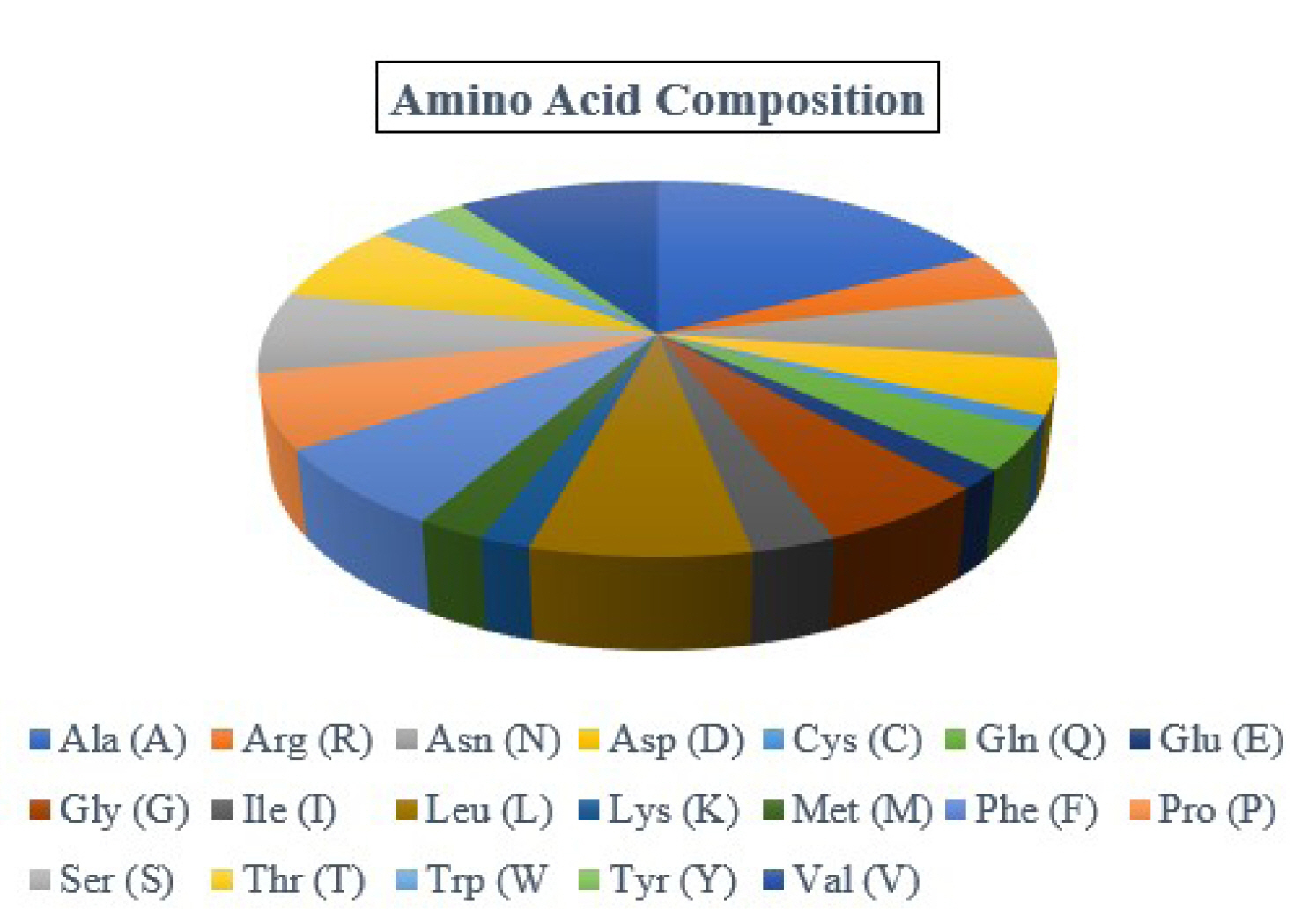
Fig.┬Ā2.
The STRING network determines the protein-protein (pr-pr) interactions. For node content: colored nodesŌĆōquery proteins and first shell of interactors, white nodesŌĆōthe second shell of interactors, empty nodesŌĆōproteins of unknown 3D structure, filled nodesŌĆōsome 3D structure is known or predicted.
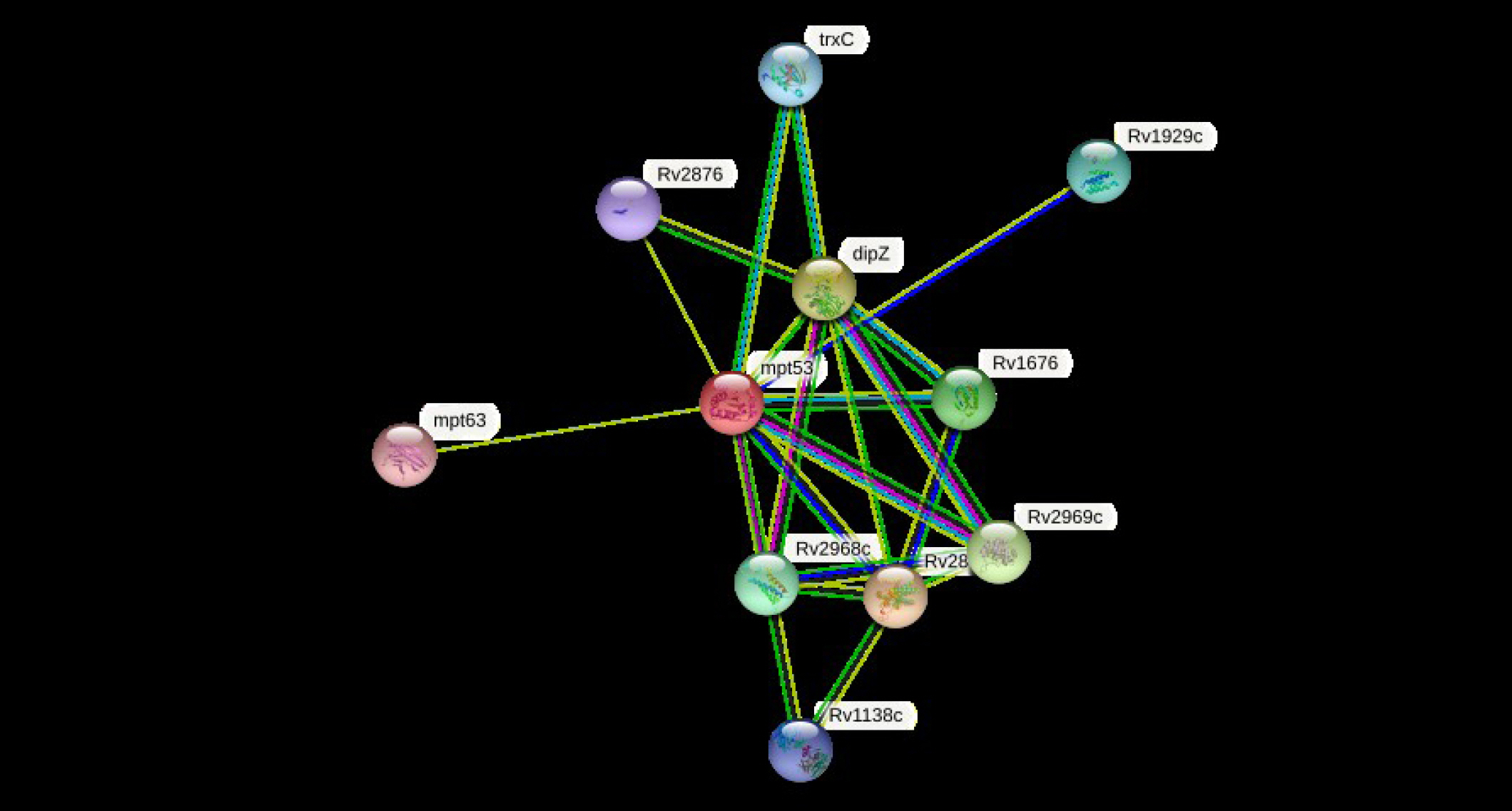
Fig.┬Ā3.
Secondary structural elements. The alpha helix (n = 66, 38.15%), extended strand (n = 39, 22.54%), beta-turn (n = 13, 7.52%), and random coil (n = 55, 31.79%).
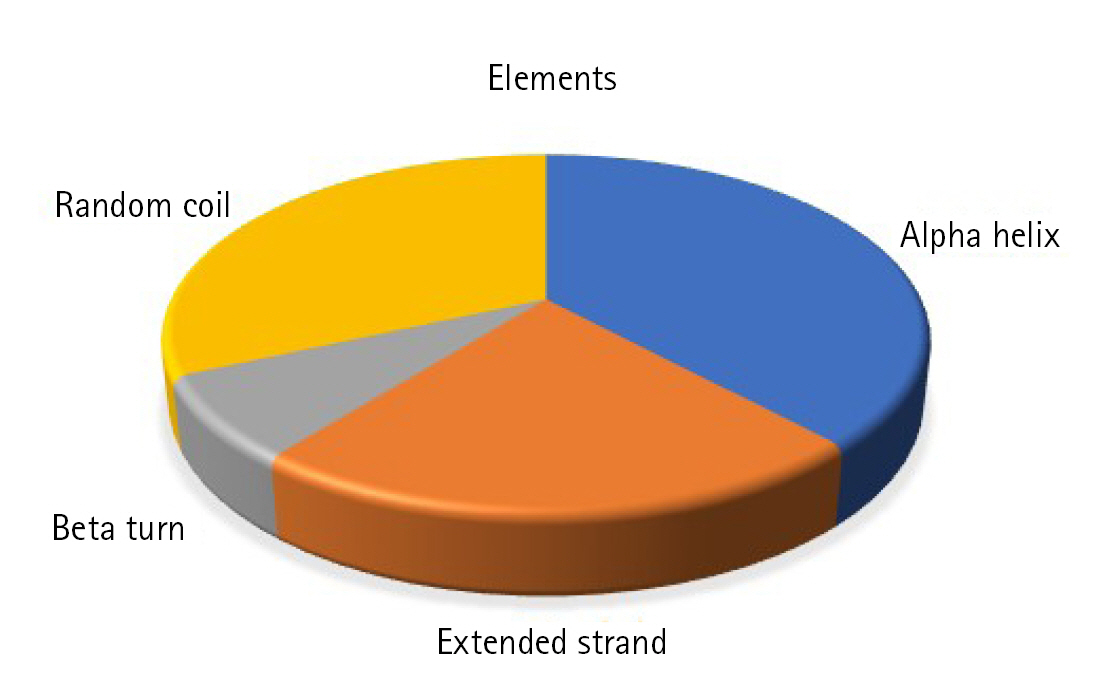
Fig.┬Ā4.
The secondary structural inquiry and assessments. (A) The amino acid types. (B) Sequence plot. (C) Secondary structure.

Fig.┬Ā5.
Tertiary structural assessment. (A) The 3D structure anticipated by Modeller. (B) The 3D structural assessment by Ramachandran plot statistics obtained from the SAVES program. The residues in most favored regions (n = 106, 91.4%), residues in additional allowed regions (n = 10, 8.6%), the number of glycine residues (shown as triangles) is 8, and the number of proline residues is 9. (C) The Z-score (ŌĆō6.53) to assess the 3D structure.
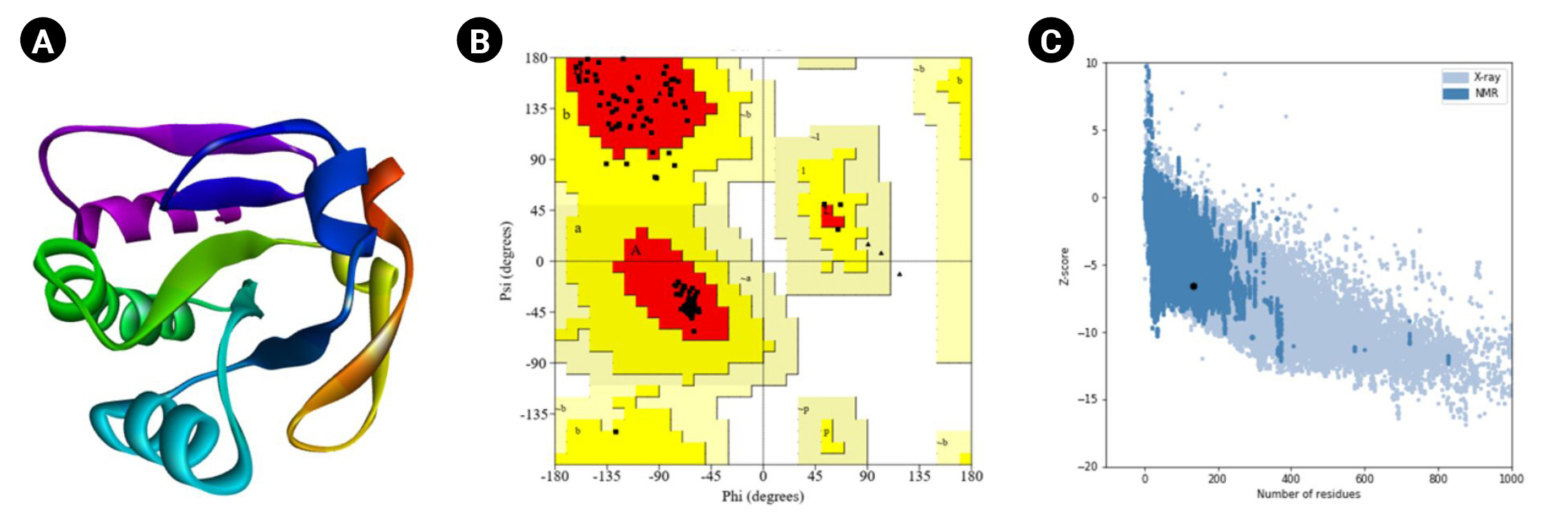
Fig.┬Ā6.
Active site determination. (A) Active sites of the protein. The ŌĆ£red sphereŌĆØ indicates the active sites of the protein. (B) The amino acid residues in the active site (blue color).
Table┬Ā1.
Protein retrieval
Table┬Ā2.
Physicochemical parameters
| Parameter | Value |
|---|---|
| Molecular weight | 18,382.98 |
| Formula | C835H1274N218O239S6 |
| Theoretical pI | 5.19, 4.98a |
| Total number of atoms | 2,572 |
| Total number of positively charged residues (Arg + Lys) | 10 |
| Total number of negatively charged residues (Asp + Glu) | 11 |
| The estimated half-life | a)┬Ā 30 h (mammalian reticulocytes, in vitro) |
| b)┬Ā >20 h (yeast, in vivo) | |
| c)┬Ā >10 h (Escherichia coli, in vivo) | |
| Aliphatic index | 86.42 |
| Instability index (II) | 29.40 |
| Grand average of hydropathicity (GRAVY) | 0.334 |
References
1. Aitken ML, Limaye A, Pottinger P, Whimbey E, Goss CH, Tonelli MR, et al. Respiratory outbreak of Mycobacterium abscessus subspecies massiliense in a lung transplant and cystic fibrosis center. Am J Respir Crit Care Med 2012;185:231ŌĆō232.


2. Pawlik A, Garnier G, Orgeur M, Tong P, Lohan A, Le Chevalier F, et al. Identification and characterization of the genetic changes responsible for the characteristic smooth-to-rough morphotype alterations of clinically persistent Mycobacterium abscessus. Mol Microbiol 2013;90:612ŌĆō629.


3. Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, et al. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 2013;381:1551ŌĆō1560.



4. Gordon SV, Bottai D, Simeone R, Stinear TP, Brosch R. Pathogenicity in the tubercle bacillus: molecular and evolutionary determinants. Bioessays 2009;31:378ŌĆō388.


5. Springer B, Stockman L, Teschner K, Roberts GD, Bottger EC. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J Clin Microbiol 1996;34:296ŌĆō303.




6. Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res 2008;18:729ŌĆō741.



7. Kaur D, Guerin ME, Skovierova H, Brennan PJ, Jackson M. Chapter 2: Biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Adv Appl Microbiol 2009;69:23ŌĆō78.



8. Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev 2003;16:463ŌĆō496.




9. Shang S, Gibbs S, Henao-Tamayo M, Shanley CA, McDonnell G, Duarte RS, et al. Increased virulence of an epidemic strain of Mycobacterium massiliense in mice. PLoS One 2011;6:e24726.



11. Gutierrez MC, Brisse S, Brosch R, Fabre M, Omais B, Marmiesse M, et al. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog 2005;1:e5.



12. Sambou T, Dinadayala P, Stadthagen G, Barilone N, Bordat Y, Constant P, et al. Capsular glucan and intracellular glycogen of Mycobacterium tuberculosis: biosynthesis and impact on the persistence in mice. Mol Microbiol 2008;70:762ŌĆō774.



13. Sani M, Houben EN, Geurtsen J, Pierson J, de Punder K, van Zon M, et al. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog 2010;6:e1000794.



14. Dhanuka R, Singh JP. Protein function prediction using functional inter-relationship. Comput Biol Chem 2021;95:107593.


15. Gabaldon T, Huynen MA. Prediction of protein function and pathways in the genome era. Cell Mol Life Sci 2004;61:930ŌĆō944.




16. Rost B, Liu J, Nair R, Wrzeszczynski KO, Ofran Y. Automatic prediction of protein function. Cell Mol Life Sci 2003;60:2637ŌĆō2650.




17. dos Santos GC, dos Santos DM, Olivieri JR, Canduri F, Silva RG, et al. Docking and small angle X-ray scattering studies of purine nucleoside phosphorylase. Biochem Biophys Res Commun 2003;309:923ŌĆō928.


18. Mills CL, Beuning PJ, Ondrechen MJ. Biochemical functional predictions for protein structures of unknown or uncertain function. Comput Struct Biotechnol J 2015;13:182ŌĆō191.



19. de Azevedo WF. Molecular dynamics simulations of protein targets identified in Mycobacterium tuberculosis. Curr Med Chem 2011;18:1353ŌĆō1366.


20. Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, et al. Protein identification and analysis tools on the ExPASy server. In: The Proteomics Protocols Handbook (Walker JM, ed.). Totowa: Humana Press, 2005. pp. 571-607.
21. Yu CS, Lin CJ, Hwang JK. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci 2004;13:1402ŌĆō1406.



22. Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins 2006;64:643ŌĆō651.


23. Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010;26:1608ŌĆō1615.




24. Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol 1998;283:489ŌĆō506.


25. Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics 2001;17:849ŌĆō850.



26. Moller S, Croning MD, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 2001;17:646ŌĆō653.



27. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001;305:567ŌĆō580.


28. Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 2017;45:D200ŌĆōD203.



29. Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res 2002;30:42ŌĆō46.



30. Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res 2014;42:D222ŌĆōD230.



31. Wilson D, Madera M, Vogel C, Chothia C, Gough J. The SUPERFAMILY database in 2007: families and functions. Nucleic Acids Res 2007;35:D308ŌĆōD313.



32. Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, et al. New and continuing developments at PROSITE. Nucleic Acids Res 2013;41:D344ŌĆōD347.



33. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607ŌĆōD613.



34. Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci 2000;25:147ŌĆō150.


35. Jones DT, Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 2015;31:857ŌĆō863.




36. Buchan DW, Jones DT. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res 2019;47:W402ŌĆōW407.




37. Bitencourt-Ferreira G, de Azevedo WF Jr. Homology modeling of protein targets with MODELLER. Methods Mol Biol 2019;2053:231ŌĆō249.


38. Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, et al. A Completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 2018;430:2237ŌĆō2243.


39. Biegert A, Mayer C, Remmert M, Soding J, Lupas AN. The MPI bioinformatics toolkit for protein sequence analysis. Nucleic Acids Res 2006;34:W335ŌĆōW339.



40. Gabler F, Nam SZ, Till S, Mirdita M, Steinegger M, Soding J, et al. Protein sequence analysis using the MPI bioinformatics toolkit. Curr Protoc Bioinformatics 2020;72:e108.



41. Bowie JU, Luthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science 1991;253:164ŌĆō170.


42. Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 2007;35:W407ŌĆōW410.



43. Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res 2018;46:W363ŌĆōW367.



44. Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics 2007;8:4.




45. Dimitrov I, Bangov I, Flower DR, Doytchinova I. AllerTOP v.2: a server for in silico prediction of allergens. J Mol Model 2014;20:2278.



46. Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R; Open Source Drug Discovery Consortium, et al. In silico approach for predicting toxicity of peptides and proteins. PLoS One 2013;8:e73957.



48. Smialowski P, Martin-Galiano AJ, Mikolajka A, Girschick T, Holak TA, Frishman D. Protein solubility: sequence based prediction and experimental verification. Bioinformatics 2007;23:2536ŌĆō2542.



49. Restrepo-Montoya D, Vizcaino C, Nino LF, Ocampo M, Patarroyo ME, Patarroyo MA. Validating subcellular localization prediction tools with mycobacterial proteins. BMC Bioinformatics 2009;10:134.




50. Saikat AS, Uddin ME, Ahmad T, Mahmud S, Imran MA, Ahmed S, et al. Structural and functional elucidation of IF-3 protein of Chloroflexus aurantiacus involved in protein biosynthesis: an in silico approach. Biomed Res Int 2021;2021:9050026.



51. Schneider G, Fechner U. Advances in the prediction of protein targeting signals. Proteomics 2004;4:1571ŌĆō1580.


52. Saikat AS. An in silico approach for potential natural compounds as inhibitors of protein CDK1/Cks2. Chem Proc 2021;8:5.

53. Bagos PG, Liakopoulos TD, Hamodrakas SJ. Evaluation of methods for predicting the topology of beta-barrel outer membrane proteins and a consensus prediction method. BMC Bioinformatics 2005;6:7.



54. Ren Z, Ren PX, Balusu R, Yang X. Transmembrane helices tilt, bend, slide, torque, and unwind between functional states of rhodopsin. Sci Rep 2016;6:34129.




55. Wong WC, Maurer-Stroh S, Schneider G, Eisenhaber F. Transmembrane helix: simple or complex. Nucleic Acids Res 2012;40:W370ŌĆōW375.



56. Bianchi F, Textor J, van den Bogaart G. Transmembrane helices are an overlooked source of major histocompatibility complex class I epitopes. Front Immunol 2017;8:1118.



57. Walther TH, Ulrich AS. Transmembrane helix assembly and the role of salt bridges. Curr Opin Struct Biol 2014;27:63ŌĆō68.


58. Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res 2015;43:D222ŌĆōD226.



59. Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 2011;39:D225ŌĆōD229.



60. Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res 2004;32:W327ŌĆōW331.



61. Goulding CW, Apostol MI, Gleiter S, Parseghian A, Bardwell J, Gennaro M, et al. Gram-positive DsbE proteins function differently from Gram-negative DsbE homologs: a structure to function analysis of DsbE from Mycobacterium tuberculosis. J Biol Chem 2004;279:3516ŌĆō3524.


62. Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer EL, et al. Pfam: the protein families database in 2021. Nucleic Acids Res 2021;49:D412ŌĆōD419.




64. Gleason FK, Holmgren A. Thioredoxin and related proteins in procaryotes. FEMS Microbiol Rev 1988;4:271ŌĆō297.


66. Eklund H, Gleason FK, Holmgren A. Structural and functional relations among thioredoxins of different species. Proteins 1991;11:13ŌĆō28.


67. Zeller T, Klug G. Thioredoxins in bacteria: functions in oxidative stress response and regulation of thioredoxin genes. Naturwissenschaften 2006;93:259ŌĆō266.



68. Kim MK, Zhao L, Jeong S, Zhang J, Jung JH, Seo HS, et al. Structural and biochemical characterization of thioredoxin-2 from Deinococcus radiodurans. Antioxidants (Basel) 2021;10:1843.



69. Marsh JA, Teichmann SA. Structure, dynamics, assembly, and evolution of protein complexes. Annu Rev Biochem 2015;84:551ŌĆō575.


70. Sowmya G, Ranganathan S. Protein-protein interactions and prediction: a comprehensive overview. Protein Pept Lett 2014;21:779ŌĆō789.


71. Xie L, Bourne PE. Functional coverage of the human genome by existing structures, structural genomics targets, and homology models. PLoS Comput Biol 2005;1:e31.



72. Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, von Mering C. Version 4.0 of PaxDb: protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics 2015;15:3163ŌĆō3168.




73. Chim N, Riley R, The J, Im S, Segelke B, Lekin T, et al. An extracellular disulfide bond forming protein (DsbF) from Mycobacterium tuberculosis: structural, biochemical, and gene expression analysis. J Mol Biol 2010;396:1211ŌĆō1226.



74. Premkumar L, Heras B, Duprez W, Walden P, Halili M, Kurth F, et al. Rv2969c, essential for optimal growth in Mycobacterium tuberculosis, is a DsbA-like enzyme that interacts with VKOR-derived peptides and has atypical features of DsbA-like disulfide oxidases. Acta Crystallogr D Biol Crystallogr 2013;69:1981ŌĆō1994.



75. Nagai S, Wiker HG, Harboe M, Kinomoto M. Isolation and partial characterization of major protein antigens in the culture fluid of Mycobacterium tuberculosis. Infect Immun 1991;59:372ŌĆō382.




76. Hahn MY, Raman S, Anaya M, Husson RN. The Mycobacterium tuberculosis extracytoplasmic-function sigma factor SigL regulates polyketide synthases and secreted or membrane proteins and is required for virulence. J Bacteriol 2005;187:7062ŌĆō7071.




77. Chim N, Harmston CA, Guzman DJ, Goulding CW. Structural and biochemical characterization of the essential DsbA-like disulfide bond forming protein from Mycobacterium tuberculosis. BMC Struct Biol 2013;13:23.




78. Hodge EA, Benhaim MA, Lee KK. Bridging protein structure, dynamics, and function using hydrogen/deuterium-exchange mass spectrometry. Protein Sci 2020;29:843ŌĆō855.




79. Uversky VN. Protein intrinsic disorder and structure-function continuum. Prog Mol Biol Transl Sci 2019;166:1ŌĆō17.


80. Hu J, Han J, Li H, Zhang X, Liu LL, Chen F, et al. Human embryonic kidney 293 cells: a vehicle for biopharmaceutical manufacturing, structural biology, and electrophysiology. Cells Tissues Organs 2018;205:1ŌĆō8.



81. Liu ZP, Wu LY, Wang Y, Zhang XS, Chen L. Bridging protein local structures and protein functions. Amino Acids 2008;35:627ŌĆō650.




82. Hegyi H, Gerstein M. The relationship between protein structure and function: a comprehensive survey with application to the yeast genome. J Mol Biol 1999;288:147ŌĆō164.


83. Li WG, Xu TL. Emerging approaches to probing ion channel structure and function. Neurosci Bull 2012;28:351ŌĆō374.




84. Lobb B, Doxey AC. Novel function discovery through sequence and structural data mining. Curr Opin Struct Biol 2016;38:53ŌĆō61.


85. Hameduh T, Haddad Y, Adam V, Heger Z. Homology modeling in the time of collective and artificial intelligence. Comput Struct Biotechnol J 2020;18:3494ŌĆō3506.



86. Saikat AS, Islam R, Mahmud S, Imran MA, Alam MS, Masud MH, et al. Structural and functional annotation of uncharacterized protein NCGM946K2_146 of Mycobacterium tuberculosis: an in-silico approach. Proceedings 2020;66:13.

87. Alva V, Nam SZ, Soding J, Lupas AN. The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res 2016;44:W410ŌĆōW415.



88. Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature 1992;356:83ŌĆō85.



89. Conklin L, Hviid A, Orenstein WA, Pollard AJ, Wharton M, Zuber P. Vaccine safety issues at the turn of the 21st century. BMJ Glob Health 2021;6(Suppl 2):e004898.



90. Riese P, Schulze K, Ebensen T, Prochnow B, Guzman CA. Vaccine adjuvants: key tools for innovative vaccine design. Curr Top Med Chem 2013;13:2562ŌĆō2580.


91. De Groot AS, Moise L, Terry F, Gutierrez AH, Hindocha P, Richard G, et al. Better epitope discovery, precision immune engineering, and accelerated vaccine design using immunoinformatics tools. Front Immunol 2020;11:442.



92. Liljeroos L, Malito E, Ferlenghi I, Bottomley MJ. Structural and computational biology in the design of immunogenic vaccine antigens. J Immunol Res 2015;2015:156241.




93. He L, Zhu J. Computational tools for epitope vaccine design and evaluation. Curr Opin Virol 2015;11:103ŌĆō112.



94. Crowcroft NS, Klein NP. A framework for research on vaccine effectiveness. Vaccine 2018;36:7286ŌĆō7293.


95. Vela Ramirez JE, Sharpe LA, Peppas NA. Current state and challenges in developing oral vaccines. Adv Drug Deliv Rev 2017;114:116ŌĆō131.



96. Wallis J, Shenton DP, Carlisle RC. Novel approaches for the design, delivery and administration of vaccine technologies. Clin Exp Immunol 2019;196:189ŌĆō204.




97. Wang J, Yu Y, Zhao Y, Zhang D, Li J. Evaluation and integration of existing methods for computational prediction of allergens. BMC Bioinformatics 2013;14 Suppl 4:S1.




98. Dimitrov I, Flower DR, Doytchinova I. AllerTOP: a server for in silico prediction of allergens. BMC Bioinformatics 2013;14 Suppl 6:S4.




99. Duan X, Zhang M, Chen F. Prediction and analysis of antimicrobial peptides from rapeseed protein using in silico approach. J Food Biochem 2021;45:e13598.



- TOOLS
-
METRICS

-
- 1 Crossref
- 0 Scopus
- 2,854 View
- 98 Download
- Related articles in GNI







