 |
 |
- Search
| Genomics Inform > Volume 21(3); 2023 > Article |
|
Abstract
Preterm birth (PTB), a pregnancy-related disease, is defined as a birth before 37 weeks of gestation. It is a major cause of maternal mortality and morbidity worldwide, and its incidence rate is steadily increasing. Various genetic factors can contribute to the etiology of PTB. Vascular endothelial growth factor A (VEGFA) gene is an important angiogenic gene and its polymorphisms have been reported to be associated with PTB development. Therefore, we conducted a case-control study to evaluate the association between VEGFA rs699947, rs2010963, and rs3025039 polymorphisms and PTB in Korean women. A total of 271 subjects (116 patients with PTB and 155 women at Ōēź38 weeks of gestation) were analyzed in this study. The genotyping of VEGFA gene polymorphisms was performed using polymerase chain reactionŌĆōrestriction fragment length polymorphism. No significant association between the patients with PTB and the control groups was confirmed. In the combination analysis, we found a significant association between PTB and VEGFA rs699947 CC-rs2010963 GG-rs3025039 CC combination (odds ratio, 3.77; 95% confidence interval, 1.091 to 13.032; p = 0.031). The VEGFA rs699947, rs2010963, and rs3025039 polymorphisms might have no genetic association with the pathogenesis of PTB in Korean women. However, the combination analysis indicates the possibility that VEGFA acts in PTB pathophysiology. Therefore, larger sample sets and replication studies are required to further elucidate our findings.
Preterm birth (PTB) refers to pregnancy-related disorders that are delivered before 37 weeks of gestation [1,2]. PTB is largely responsible for the deaths of children [3]. According to a previous study, approximately 15 million babies are born preterm yearly, and the prevalence rate is approximately 5%ŌĆō18% [4]. In South Korea, PTB accounted for 7.2% of births in 2016, a 1.5-fold increase from 2006 [5]. However, the exact causes of PTB remain unclear [6].
Various studies have shown that PTB is a syndrome caused by multiple mechanisms including inflammatory infection and uteroplacental ischemia (UPI) [7-10]. Particularly, the causes of UPI are mostly due to the abnormal pattern of the placental stem artery development, accompanied by an insufficient increase in capillary angiogenesis, and terminal villous dysplasia [11]. The main pathways involved in vascular dysplasia are vascular endothelial growth factor (VEGF), angiopoietin-TIE2, and transforming growth factor-╬▓ [12]. Among them, the VEGF family-mediated angiogenesis plays a key role [13-16]. VEGF is an angiogenic factor that also participates in the formation of blood vessels during the development of the corpus luteum [17]. Additionally, a previous study reported on the study of in vivo model systems showed that VEGF could regulate circulating progesterone levels [18]. Moreover, blocking of VEGF second-trimester signal causes impairment of luteal circulation and induces PTB [19,20]. Also, the American College of Obstetricians and Gynecologists suggested that the progesterone supplement is a useful way to reduce the risk of PTB [21].
In the VEGF family, vascular endothelial growth factor A (VEGFA) is an angiogenic factor for developing the embryonic vasculature [22]. Related studies of maternal serum levels found that an increase in VEGFA would promote PTB development [23,24]. Additionally, the VEGFA polymorphisms have been reported to be associated with altered VEGFA protein expression [25,26]. VEGFA is located on chromosome 6 (6p12) [27]. Papazoglou et al. [28] confirmed the association between rs2010963 in VEGFA and spontaneous preterm births (sPTB). Also, a previous study has shown that VEGFA rs699947 polymorphism is associated with the risk of sPTB [20]. A genetic-environmental study also confirmed that rs3025039 is associated with the risk of PTB [29]. However, to the best of our knowledge, a genetic association study between VEGFA and PTB has not been conducted on Korean women. Therefore, we investigated the genetic correlation between the three VEGFA gene polymorphisms (rs699947, rs2010963, and rs3025039) and PTB in Korean women. We also analyzed the correlation of three polymorphisms of VEGFA with characteristic data.
We analyzed a total of 271 samples that include 116 patients with PTB and 155 control groups recruited by the Department of Obstetrics and Gynecology of Dankook University Hospital in Korea (Table 1). In the screening of experimental subjects, PTB patients were composed of women who gave birth before 37 weeks, and the control groups were composed of women who gave birth after 37 weeks. The patients with PTB and the control groups did not include women with multiple pregnancy, gestational diabetes mellitus, pre-gestational hypertension, and early placental separation. The study was conducted after approval by the Institutional Review Board of Dankook University Hospital F (date of approval and the project identification code are respectively 16 November 2017 and DKUH 2016-12-003-005).
DNA was extracted from peripheral blood or buccal cells using the GeneAll Exgene Clinic SV mini kit (GeneAll, Seoul, Korea). Genotypes of VEGFA rs699947, rs2010963, and rs3025039 polymorphisms were detected using the polymerase chain reactionŌĆōrestriction fragment length polymorphism. Primer sets were designed for a determination of rs699947, rs2010963, and rs3025039 polymorphisms reported by previous studies (Table 2) [30-32]. Each polymerase chain reaction (PCR) reaction was performed in a total volume of 20ŌĆ»╬╝L mixture containing 10 ng of genomic DNA, 10 pM of each primer, 0.2 mM MgCl2, 10├Ś PCR buffer, and 1.0 U NV DNA polymerase (NAVI BioTech, Cheonan, Korea). The PCR amplifications were conducted with a C1000 Touch thermal cycler (Bio-Rad, Hercules, CA, USA) under the following conditions: 95Ōäā for 5 min, followed by 32 cycles at 95Ōäā for 30 s, 61Ōäā annealing 30 s, and 72Ōäā for 50 s and then a final extension at 72┬░C for 10 min for rs699947; 95Ōäā for 5 min, followed by 35 cycles at 94Ōäā for 1 min, 62Ōäā annealing 1 min, and 72Ōäā for 1 min and then a final extension at 72┬░C for 5 min for rs2010963; 94Ōäā for 3 min, followed by 35 cycles at 94Ōäā for 30 s, 58Ōäā annealing 30 s, and 72Ōäā for 30 s and then a final extension at 72┬░C for 10 min for rs3025039. Each of the PCR product was digested with 1.0 U BglŌģĪ (rs699947 C>A), BsmFI (rs2010963 C>G), and NlaŌģó (rs3025039 C>T) restriction enzymes (Enzynomics, Daejeon, Korea) for 6 h at 37┬░C (BglŌģĪ and NlaŌģó) and 65┬░C (BsmFI), respectively. The polymorphic BglII site for rs699947 produced 325 bp (C allele) or 202 bp and 123 bp (A allele) fragments (Fig. 1). The restricted alleles of rs2010963 were 469 bp (C allele) or 274 bp and 195 bp (G allele) fragments, and rs3025039 was confirmed by different fragment sizes of 266 bp (C allele) or 208 bp and 54 bp (T allele) (Figs. 2 and 3).
A test of cross tabulation analysis was performed using the SPSS 26 Statistics (IBM Corp., Armonk, NY, USA). The chi-squared tests were used to assess the Hardy-Weinberg equilibrium. We used SISA (Simple Interactive Statistical Analysis, https://www.quantitativeskills.com/sisa/) to compare the genotype or allele frequencies between groups through a 2 ├Ś r table and to calculate odds ratio (OR) with 95% confidence intervals (CIs) using a 2 ├Ś 2 table. Based on genotype data, linkage disequilibrium (LD) was estimated using the HaploView 4.2. Haplotype analysis was performed to compare haplotype frequencies between the patients with preterm and control groups by HAPSTAT software v.3.0. In addition, combination analyses of genotype frequency were performed using the SNPstats web-based tool (SNPstats, https://www.snpstats.net/snpstats/). A p-value of <0.05 was considered statistically significant. Bonferroni correction was applied to adjust for multiple comparisons [33].
In this study, 271 pregnant women (including 116 patients with PTB and 155 control groups) were analyzed. The mean values of age, height, weight, pre- and post-pregnancy weight, systolic blood pressure, and diastolic blood pressure were insignificantly different between the patients with PTB and control groups (p > 0.05). However, a significant difference was found in birth weight and gestational age between patients with PTB and control groups (p < 0.05) (Table 1).
Genotype and allele distributions of VEGFA gene polymorphisms for the 116 patients with PTB and 155 controls are presented in Table 2. The genotype and allele frequencies of rs699947, rs2010963, and rs3025039 were insignificantly associated with PTB (p > 0.05) (Tables 3 and 4).
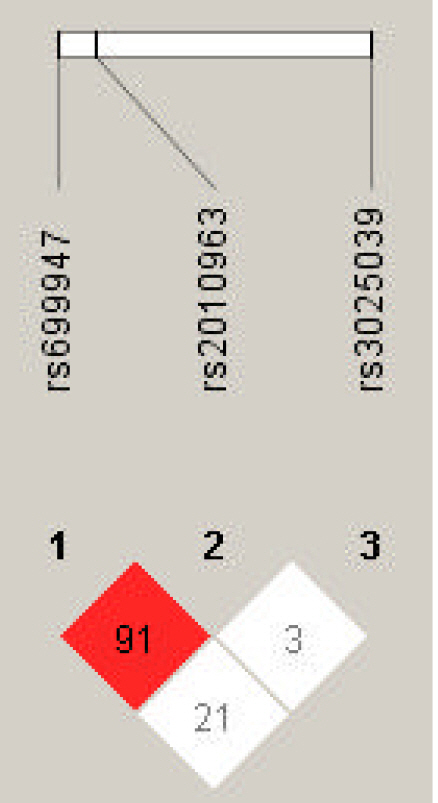
LD analysis was performed for rs699947, rs2010963, and rs3025039 polymorphisms in patients with PTB and controls. The LD between rs699947 and rs2010963 was high (DŌĆÖ = 0.91, r2 = 1.0) (Fig. 4). Therefore, we analyzed the frequencies of the four common haplotypes. No significant difference was found between the rs699947 and rs2010963 polymorphism and PTB incidence (p > 0.05) (Supplementary Table 1).
In the genotype combination analysis of the three gene polymorphisms (rs699947 C>A, rs2010963 C>G, and rs3025039 C>T), we found a significant association with PTB from rs699947 CC-rs2010963 GG-rs3025039 CC combination (OR, 3.77; 95% CI, 1.091 to 13.032; p = 0.031) (Table 5).
The VEGF protein levels increase during late pregnancy and peak during the time of cervical ripening [34]. It is reported that higher VEGF protein expression in the early stages could lead to premature cervical ripening, resulting in PTB [34,35]. Several studies reported that VEGFA polymorphisms are associated with altered VEGF production [25]. Among them, the rs699947 CC homozygotes and rs2010963 C allele were associated with higher secretion of VEGF [23]. Moreover, in carriers of the rs3025039 T allele, VEGF plasma levels were significantly lower [26]. Since VEGFA genetically controls VEGF protein expression [36-38]. We analyzed the genetic associations between VEGFA rs699947, rs2010963, and rs3025039 polymorphisms and PTB in Korean women.
The influence of VEGFA gene polymorphisms on PTB has been inconsistent to date, possibly because of ethnic heterogeneity [39]. In this analysis, no significant association between rs2010963 polymorphism and PTB was found (p > 0.05) (Tables 3 and 4). Contrarily, VEGFA rs2010963 G allele was significantly associated with PTB in the Malaysian population [20]. The minor allele frequency (MAF) of South Asian population is different from that of Korean population (Table 6). However, rs699947 and rs2010963 are also in a strong LD in Malaysians (DŌĆ▓ = 0.97, r2 = 1.0) [20]. Therefore, we carefully speculate that the significant association was not replicated because of the genetic heterogeneity. For evaluating the effect of these two polymorphisms in VEGFA for PTB, further studies need to be examined in different populations. Also, in the results of VEGFA rs699947 and rs3025039, no significant association in the Korean population (p > 0.05) (Table 3 and 4). This is consistent with the findings of Langmia et al. [20], which suggested the VEGFA rs699947 and rs3025039 were insignificantly associated with PTB in the Malaysian population. Contrastively, the rs699947 AA genotype was associated with a risk of PTB in Australian and New Zealand populations [29]. Also, rs3025039 CT and TT genotype of VEGFA was associated with an increased risk of sPTB in the Greek population [28]. MAFs for each VEGFA polymorphism were different in Korea population (in East Asia population), Europe, and South Asia population (Table 6). Also, each population is genetically distinct [40]. It can be seen that the effect of VEGFA gene on PTB is not consistent in different populations, which may be due to the genetic heterogeneity [39]. Zhao et al. [29] reported an increased risk of PTB in women with the VEGFA rs3025039 CC genotype among the women exposed to higher air quality index levels. Although the study considered the influence of environmental factors, it also confirmed that rs3025039 was potentially associated with PTB pathogenesis [29]. Therefore, more studies in other populations are needed to determine the association of VEGFA gene polymorphisms with PTB development.
Haplotype analysis provides more information about identifying key candidate genes for PTB than testing a single locus, and it reduces the false-positive associations in the result [41,42]. Hence, we performed the haplotype analysis of VEGFA rs2010963, rs699947, and rs3025039 by LD analysis (Fig. 4). The LD between rs2010963 and rs699947 was high (DŌĆ▓ = 0.91). The rs699947 CC-rs2010963 GG combination showed a marginal trend toward significance (OR, 0.36; 95% CI, 0.125 to 1.021; p = 0.051). However, no significant result in the haplotype analysis was found (Supplementary Table 1). In contrast, in the genotype combination analysis, the rs699947 CC-rs2010963 GG-rs3025039 CC combination was associated significantly with patients with PTB (OR, 3.77; 95% CI, 1.091 to 13.032; p = 0.031) (Table 5). According to this result, we may speculate that the VEGFA rs699947 CC-rs2010963 GG-rs3025039 CC combination plays an etiological role in PTB development in Korean women. However, the significant results were not maintained after correction for multiple testing. Therefore, VEGFA polymorphisms, which were analyzed in this study, were not associated with PTB in Korean women.
This study has several limitations. Firstly, the sample size used in this study was relatively small. However, in the statistical analysis using the G*Power program, sample power of over 90% was obtained in 271 subjects, exceeding the minimum limit (80%) for clinical studies [43]. Nonetheless, more samples would be requested to validate our results. Secondly, only one gene was investigated in this study, and gene-gene or gene-environment interactions were not considered. PTB is generally not caused by the influence of a single therapeutic gene or an environmental factor [44]. Thus, it was concluded that VEGFA and other genes work together to cause PTB. Finally, this study included only Korean women. Since we do not replicate a significant association between polymorphisms in VEGFA and PTB, further studies conducted in various populations will be required. Research of the wider populations can reveal missed insights into the association between genetic markers and phenotype [45]. Analysis in other populations is critical to assess the accuracy and wider relevance of a finding. Hence, this study is limited to Korea and may need to be replicated in other countries, including different populations allowing assessment of potential racial or ethnic heterogeneity.
Despite these limitations, this study is the first analysis of the association between VEGFA gene polymorphisms and PTB in Korean women. Conclusively, our study could not confirm genetic relationships between the VEGFA rs699947, rs2010963, and rs3025039 polymorphisms, and PTB pathogenesis. Therefore, current evidence does not support using the VEGFA to predict risk or as a marker for therapeutic intervention in Korea. Replication studies with larger sample sizes and different populations are still needed to elucidate the genetic relationship between the VEGFA gene polymorphisms and the pathogenesis of PTB.
Notes
Acknowledgments
We are grateful to all volunteers for providing DNA samples. This work was supported by the National Research Foundation of Korea Grant funded by the Korea Government (NRF-2019R1D1A3A03103804).
Supplementary Materials
Supplementary data can be found with this article online at http://www.genominfo.org.
Supplementary┬ĀTable┬Ā1.
Haplotype frequencies of VEGFA polymorphisms in the patients with preterm birth and control group
Fig.┬Ā1.
The polymerase chain reactionŌĆōrestriction fragment length polymorphism result of vascular endothelial growth factor A (VEGFA) rs699947 polymorphism. Lanes are as follows: M, 50 bp DNA size ladder; 1, 2, 4, 5, CC genotype; 3, AA genotype; 6, 7, CA genotype.
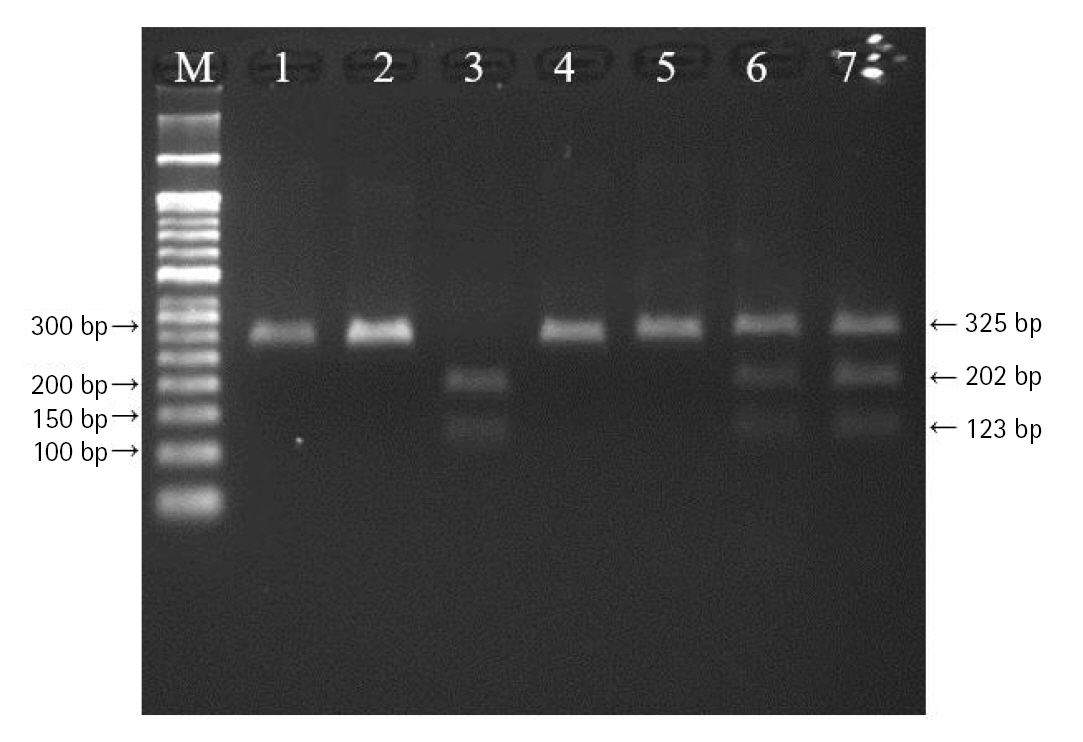
Fig.┬Ā2.
The polymerase chain reactionŌĆōrestriction fragment length polymorphism result of vascular endothelial growth factor A (VEGFA) rs2010963 polymorphism. Lanes are as follows: M, 50 bp DNA size ladder; 1, CC genotype; 2, 3, 6, 7, GC genotype; 4, 5, GG genotype.

Fig.┬Ā3.
The polymerase chain reactionŌĆōrestriction fragment length polymorphism result of vascular endothelial growth factor A (VEGFA) rs3025039 polymorphism. Lanes are as follows: M, 50 bp DNA size ladder; 1, 4, 5, CC genotype; 3, 6, CT genotype; 2, 7, TT genotype.
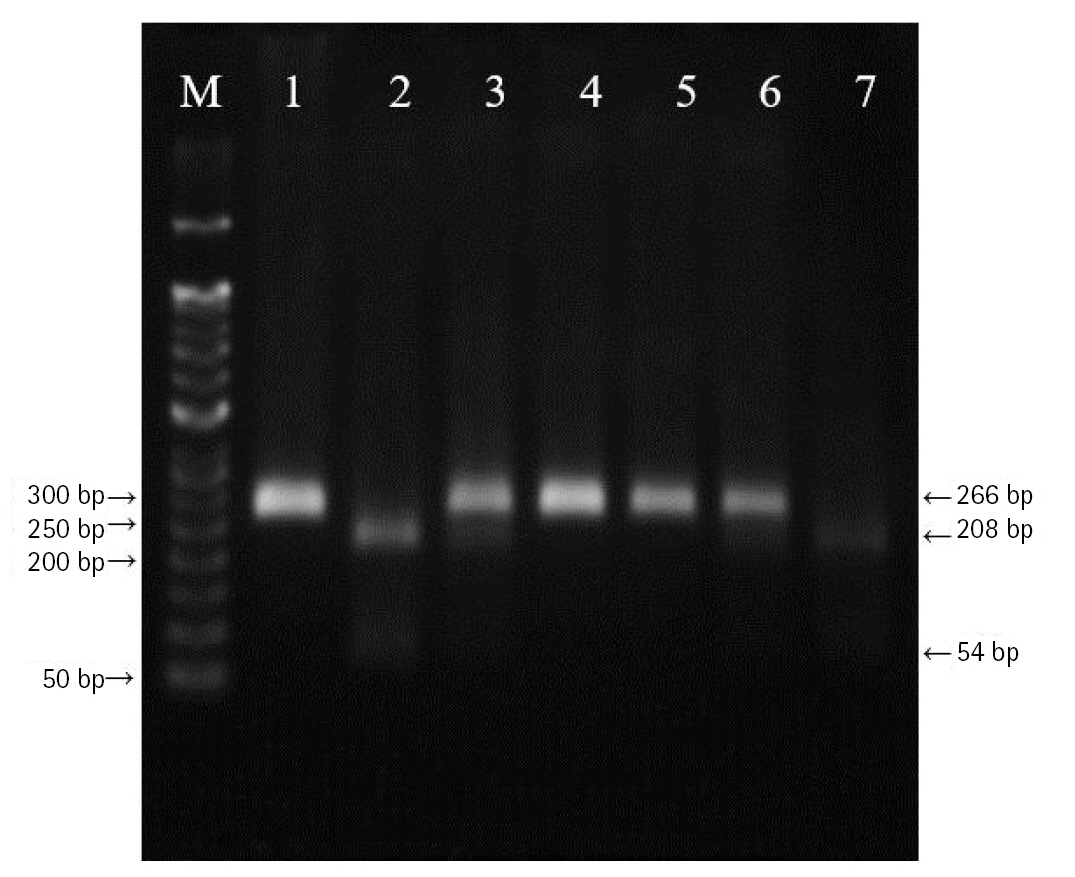
Fig.┬Ā4.
Linkage disequilibrium (LD) plot of single nucleotide polymorphisms of vascular endothelial growth factor A (VEGFA). Numbers represent LD between markers based on the DŌĆ▓ values.
Table┬Ā1.
Characteristics of patients with preterm birth and control groups
| Characteristic | Preterm birth (n=116) | Control (n=155) | p-valuea |
|---|---|---|---|
| Age (y) | 30.59 ┬▒ 4.59 | 30.41 ┬▒ 4.91 | 0.779 |
| Heigh (cm) | 160.29 ┬▒ 4.59 | 160.56 ┬▒ 5.56 | 0.697 |
| Pre-pregnant weight (g) | 56.82 ┬▒ 9.92 | 56.1 ┬▒ 10.41 | 0.599 |
| Post-pregnant weight (g) | 67.37 ┬▒ 10.51 | 69.46 ┬▒ 10.45 | 0.140 |
| SBP (mmHg) | 120.24 ┬▒ 14.97 | 121.32 ┬▒ 14.47 | 0.585 |
| DBP (mmHg) | 74.76 ┬▒ 12.82 | 75.79 ┬▒ 11.87 | 0.535 |
| Birth weight (g) | 2,354.51 ┬▒ 643.04 | 3,158.46 ┬▒ 463.68 | < 0.001* |
| GA (wk) | 33.19 ┬▒ 3.06 | 38.74 ┬▒ 1.06 | < 0.001* |
Table┬Ā2.
PCR primer sequences, annealing temperature and the expected sizes of PCR products
Table┬Ā3.
Genotype and allele frequencies distribution of VEGFA gene polymorphisms in patients with preterm birth and control group
| Maker |
Genotype/allele distribution, n (%) |
||||
|---|---|---|---|---|---|
| Preterm birth (n = 116) | Control (n = 155) | p-valuea | |||
| VEGFA rs699947 | Genotype | CC | 67 (57.8) | 78 (50.3) | 0.366 |
| CA | 41 (35.3) | 68 (43.9) | |||
| AA | 8 (6.9) | 9 (7.8) | |||
| HWE | 0.617 | 0.240 | |||
| Allele | C | 175 (75.4) | 242 (73.8) | 0.407 | |
| A | 57 (24.6) | 86 (26.2) | |||
| rs2010963 | Genotype | CC | 24 (20.7) | 32 (20.6) | 0.155 |
| CG | 51 (44.0) | 84 (54.2) | |||
| GG | 41 (35.3) | 39 (25.2) | |||
| HWE | 0.275 | 0.284 | |||
| Allele | C | 99 (42.7) | 148 (47.7) | 0.241 | |
| G | 133 (57.3) | 162 (52.3) | |||
| rs3025039 | Genotype | CC | 81 (69.8) | 102 (65.8) | 0.490 |
| CT | 31 (26.7) | 50 (32.3) | |||
| TT | 4 (3.4) | 3 (1.9) | |||
| HWE | 0.632 | 0.264 | |||
| Allele | C | 193 (83.2) | 254 (81.9) | 0.704 | |
| T | 39 (16.8) | 56 (36.1) | |||
Table┬Ā4.
The OR of VEGFA gene polymorphisms in patients with preterm birth and control group
| Maker |
Preterm birth vs. control |
||
|---|---|---|---|
| OR | 95% CI | p-valuea | |
| VEGFA rs699947 | |||
| ŌĆāGenotype | |||
| ŌĆāŌĆāCC | 1.00 | - | Reference |
| ŌĆāŌĆāCA | 0.70 | 0.423ŌĆō1.165 | 0.170 |
| ŌĆāŌĆāAA | 0.97 | 0.353ŌĆō2.645 | 1.000 |
| ŌĆāAllele | |||
| ŌĆāŌĆāC/A | 0.92 | 0.622ŌĆō1.350 | 0.407 |
| ŌĆāŌĆāDominant (CC/CA + AA) | 0.74 | 0.456ŌĆō1.203 | 0.225 |
| ŌĆāŌĆāRecessive (CC + CA/AA) | 0.83 | 0.311ŌĆō2.227 | 0.714 |
| ŌĆāŌĆāOverdominant (CC + AA/CA) | 0.66 | 0.409ŌĆō1.076 | 0.157 |
| rs2010963 | |||
| ŌĆāGenotype | |||
| ŌĆāŌĆāCC | 1.00 | - | Reference |
| ŌĆāŌĆāCG | 0.81 | 0.430ŌĆō1.525 | 0.513 |
| ŌĆāŌĆāGG | 1.40 | 0.705ŌĆō2.787 | 0.335 |
| ŌĆāAllele | |||
| ŌĆāŌĆāC/G | 0.82 | 0.578ŌĆō1.148 | 0.241 |
| ŌĆāŌĆāDominant (CC/CG + GG) | 1.00 | 0.550ŌĆō1.807 | 1.000 |
| ŌĆāŌĆāRecessive (CC + CG/GG) | 1.63 | 0.961ŌĆō2.751 | 0.069 |
| ŌĆāŌĆāOverdominant (CC + GG/CG) | 0.66 | 0.409ŌĆō1.076 | 0.096 |
| rs3025039 | |||
| ŌĆāGenotype | |||
| ŌĆāŌĆāCC | 1.00 | - | Reference |
| ŌĆāŌĆāCT | 0.78 | 0.457ŌĆō1.333 | 0.364 |
| ŌĆāŌĆāTT | 1.68 | 0.365ŌĆō7.716 | 0.501 |
| ŌĆāAllele | |||
| ŌĆāŌĆāC/T | 0.92 | 0.585ŌĆō0.437 | 0.704 |
| ŌĆāŌĆāDominant (CC/CT + TT) | 0.83 | 0.496ŌĆō1.395 | 0.484 |
| ŌĆāŌĆāRecessive (CC + CT/TT) | 0.76 | 0.450ŌĆō1.303 | 0.325 |
| ŌĆāŌĆāOverdominant (CC + TT/CT) | 1.81 | 0.397ŌĆō8.246 | 0.437 |
Table┬Ā5.
Combined three genotypes of VEGFA polymorphisms in the patients with preterm birth and control group
| Marker | Combination | No. (%) | p-valuea | OR (95% CI) | |
|---|---|---|---|---|---|
| Preterm birth (n = 116) | Control (n = 155) | ||||
| VEGFA rs699947 | CC - CC - CC | 14 (12.1) | 22 (14.2) | - | Reference |
| rs2010963 | CC - CC - CT | 7 (6.0) | 7 (4.5) | 0.475 | 1.57 (0.453ŌĆō5.450) |
| rs3025039 | CC - CC - TT | 2 (1.7) | 1 (0.6) | 0.347 | 3.14 (0.260ŌĆō37.992) |
| CA - CC - CC | 1 (0.9) | 1 (0.6) | 0.754 | 1.57 (0.091ŌĆō27.212) | |
| CA - CC -CT | 0 | 1 (0.6) | 0.316 | NA | |
| CA - CC - TT | 0 | 0 | 1.000 | NA | |
| AA - CC - CC | 0 | 0 | 1.000 | NA | |
| AA - CC - CT | 0 | 0 | 1.000 | NA | |
| AA - CC - TT | 0 | 0 | 1.000 | NA | |
| CC - CG - CC | 23 (19.8) | 32 (20.6) | 0.781 | 0.89 (0.375ŌĆō2.088) | |
| CC - CG - CT | 6 (5.2) | 9 (5.8) | 1.000 | 0.96 (0.279ŌĆō3.27) | |
| CC - CG - TT | 0 | 0 | 1.000 | NA | |
| CA - CG - CC | 13 (11.2) | 27 (17.4) | 0.561 | 1.32 (0.515ŌĆō3.389) | |
| CA - CG -CT | 6 (5.2) | 14 (9.0) | 0.506 | 0.67 (0.209ŌĆō2.165) | |
| CA - CG - TT | 2 (1.7) | 2 (1.3) | 0.67 | 1.57 (0.198ŌĆō12.470) | |
| AA - CG - CC | 1 (0.9) | 0 | 0.147 | NA | |
| AA - CG - CT | 0 | 0 | 1.000 | NA | |
| AA - CG - TT | 0 | 0 | 1.000 | NA | |
| CC - GG - CC | 12 (10.3) | 5 (3.2) | 0.031* | 3.77 (1.091ŌĆō13.032) | |
| CC - GG - CT | 3 (2.6) | 2 (1.3) | 0.369 | 2.36 (0.349ŌĆō15.927) | |
| CC - GG - TT | 0 | 0 | 1.000 | NA | |
| CA - GG - CC | 13 (11.2) | 13 (8.4) | 0.384 | 1.57 (0.567ŌĆō4.357) | |
| CA - GG -CT | 6 (5.2) | 10 (6.5) | 1.000 | 0.94 (0.280ŌĆō3.174) | |
| CA - GG - TT | 0 | 0 | 1.000 | NA | |
| AA - GG - CC | 4 (3.4) | 2 (1.3) | 0.203 | 3.14 (0.507ŌĆō19.492) | |
| AA - GG - CT | 3 (2.6) | 7 (4.5) | 0.606 | 0.67 (0.149ŌĆō3.047) | |
| AA - GG - TT | 0 | 0 | 1.000 | NA | |
References
1. Vogel JP, Chawanpaiboon S, Moller AB, Watananirun K, Bonet M, Lumbiganon P. The global epidemiology of preterm birth. Best Pract Res Clin Obstet Gynaecol 2018;52:3ŌĆō12.


3. Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller AB, Narwal R, et al. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet 2012;379:2162ŌĆō2172.


4. Wagura P, Wasunna A, Laving A, Wamalwa D, Ng'ang'a P. Prevalence and factors associated with preterm birth at Kenyatta National Hospital. BMC Pregnancy Childbirth 2018;18:107.




5. Kwon BN, Lee NR, Kim HJ, Kang YD, Kim JS, Park JW, et al. Folate metabolizing gene polymorphisms and genetic vulnerability to preterm birth in Korean women. Genes Genomics 2021;43:937ŌĆō945.



6. Han YS, Ha EH, Park HS, Kim YJ, Lee SS. Relationships between pregnancy outcomes, biochemical markers and pre-pregnancy body mass index. Int J Obes (Lond) 2011;35:570ŌĆō577.



7. Bastek JA, Sammel MD, Pare E, Srinivas SK, Posencheg MA, Elovitz MA. Adverse neonatal outcomes: examining the risks between preterm, late preterm, and term infants. Am J Obstet Gynecol 2008;199:367.

8. Lunde A, Melve KK, Gjessing HK, Skjaerven R, Irgens LM. Genetic and environmental influences on birth weight, birth length, head circumference, and gestational age by use of population-based parent-offspring data. Am J Epidemiol 2007;165:734ŌĆō741.


9. McIntire DD, Leveno KJ. Neonatal mortality and morbidity rates in late preterm births compared with births at term. Obstet Gynecol 2008;111:35ŌĆō41.


10. Svensson AC, Sandin S, Cnattingius S, Reilly M, Pawitan Y, Hultman CM, et al. Maternal effects for preterm birth: a genetic epidemiologic study of 630,000 families. Am J Epidemiol 2009;170:1365ŌĆō1372.


11. Todros T, Sciarrone A, Piccoli E, Guiot C, Kaufmann P, Kingdom J. Umbilical Doppler waveforms and placental villous angiogenesis in pregnancies complicated by fetal growth restriction. Obstet Gynecol 1999;93:499ŌĆō503.


12. Pang C, Lim CS, Brookes J, Tsui J, Hamilton G. Emerging importance of molecular pathogenesis of vascular malformations in clinical practice and classifications. Vasc Med 2020;25:364ŌĆō377.



13. Andraweera PH, Dekker GA, Thompson SD, Nowak RC, Zhang JV, McCowan LM, et al. Association of vascular endothelial growth factor +936 C/T single-nucleotide polymorphism with pregnancies complicated by small-for-gestational-age babies. Arch Pediatr Adolesc Med 2011;165:1123ŌĆō1130.


14. Crider KS, Whitehead N, Buus RM. Genetic variation associated with preterm birth: a HuGE review. Genet Med 2005;7:593ŌĆō604.


15. Guryanov I, Tennikova T, Urtti A. Peptide inhibitors of vascular endothelial growth factor A: current situation and perspectives. Pharmaceutics 2021;13:1337.



16. Ruhrberg C. Growing and shaping the vascular tree: multiple roles for VEGF. Bioessays 2003;25:1052ŌĆō1060.


17. Reynolds LP, Grazul-Bilska AT, Redmer DA. Angiogenesis in the corpus luteum. Endocrine 2000;12:1ŌĆō9.


18. Wada Y, Ozaki H, Abe N, Mori A, Sakamoto K, Nagamitsu T, et al. Role of vascular endothelial growth factor in maintenance of pregnancy in mice. Endocrinology 2013;154:900ŌĆō910.



19. Holmes DI, Zachary I. The vascular endothelial growth factor (VEGF) family: angiogenic factors in health and disease. Genome Biol 2005;6:209.



20. Langmia IM, Apalasamy YD, Omar SZ, Mohamed Z. Association of VEGFA gene polymorphisms and VEGFA plasma levels with spontaneous preterm birth. Pharmacogenet Genomics 2015;25:199ŌĆō204.


21. Norwitz ER. A blood test to predict preterm birth: don't mess with maternal-fetal stress. J Clin Endocrinol Metab 2009;94:1886ŌĆō1889.


22. Otrock ZK, Makarem JA, Shamseddine AI. Vascular endothelial growth factor family of ligands and receptors: review. Blood Cells Mol Dis 2007;38:258ŌĆō268.


23. Renner W, Kotschan S, Hoffmann C, Obermayer-Pietsch B, Pilger E. A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J Vasc Res 2000;37:443ŌĆō448.



24. Smith GC, Crossley JA, Aitken DA, Jenkins N, Lyall F, Cameron AD, et al. Circulating angiogenic factors in early pregnancy and the risk of preeclampsia, intrauterine growth restriction, spontaneous preterm birth, and stillbirth. Obstet Gynecol 2007;109:1316ŌĆō1324.


25. Almawi WY, Saldanha FL, Mahmood NA, Al-Zaman I, Sater MS, Mustafa FE. Relationship between VEGFA polymorphisms and serum VEGF protein levels and recurrent spontaneous miscarriage. Hum Reprod 2013;28:2628ŌĆō2635.


26. Shahbazi M, Fryer AA, Pravica V, Brogan IJ, Ramsay HM, Hutchinson IV, et al. Vascular endothelial growth factor gene polymorphisms are associated with acute renal allograft rejection. J Am Soc Nephrol 2002;13:260ŌĆō264.


27. Abhary S, Burdon KP, Gupta A, Lake S, Selva D, Petrovsky N, et al. Common sequence variation in the VEGFA gene predicts risk of diabetic retinopathy. Invest Ophthalmol Vis Sci 2009;50:5552ŌĆō5558.


28. Papazoglou D, Galazios G, Koukourakis MI, Kontomanolis EN, Maltezos E. Association of -634G/C and 936C/T polymorphisms of the vascular endothelial growth factor with spontaneous preterm delivery. Acta Obstet Gynecol Scand 2004;83:461ŌĆō465.


29. Zhao N, Wu W, Feng Y, Yang F, Han T, Guo M, et al. Polymorphisms in oxidative stress, metabolic detoxification, and immune function genes, maternal exposure to ambient air pollution, and risk of preterm birth in Taiyuan, China. Environ Res 2021;194:110659.


30. Diao LP, Yu XM, Gao YH, Li Y, Liu HS, Liu LH, et al. Association of VEGF genetic polymorphisms with the clinical characteristics of non-Hodgkin's lymphoma. J Cancer Res Clin Oncol 2009;135:1473ŌĆō1481.



31. Han SW, Kim GW, Seo JS, Kim SJ, Sa KH, Park JY, et al. VEGF gene polymorphisms and susceptibility to rheumatoid arthritis. Rheumatology (Oxford) 2004;43:1173ŌĆō1177.


32. Kim SH, Choi YM, Choung SH, Jun JK, Kim JG, Moon SY. Vascular endothelial growth factor gene +405 C/G polymorphism is associated with susceptibility to advanced stage endometriosis. Hum Reprod 2005;20:2904ŌĆō2908.


34. Mowa CN, Jesmin S, Sakuma I, Usip S, Togashi H, Yoshioka M, et al. Characterization of vascular endothelial growth factor (VEGF) in the uterine cervix over pregnancy: effects of denervation and implications for cervical ripening. J Histochem Cytochem 2004;52:1665ŌĆō1674.



35. Monangi NK, Brockway HM, House M, Zhang G, Muglia LJ. The genetics of preterm birth: progress and promise. Semin Perinatol 2015;39:574ŌĆō583.


36. Al-Khateeb GM, Mustafa FE, Sater MS, Almawi WY. Effect of the functional VEGFA-583C/T variant on vascular endothelial growth factor levels and the risk of recurrent spontaneous miscarriage. Fertil Steril 2011;95:2471ŌĆō2473.


37. Lin TH, Wang CL, Su HM, Hsu PC, Juo SH, Voon WC, et al. Functional vascular endothelial growth factor gene polymorphisms and diabetes: effect on coronary collaterals in patients with significant coronary artery disease. Clin Chim Acta 2010;411:1688ŌĆō1693.


38. Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine 2000;12:1232ŌĆō1235.


39. Crump C, Sundquist J, Sundquist K. Association of preterm birth with lipid disorders in early adulthood: a Swedish cohort study. PLoS Med 2019;16:e1002947.



40. Papageorgiou L, Andreou A, Zervou M, Vlachakis D, Goulielmos GN, Eliopoulos E. A global population genomic analysis shows novel insights into the genetic characteristics of endometriosis. World Acad Sci J 2023;5:12.

41. Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet 2001;2:91ŌĆō99.



42. Shang CY, Gau SS, Liu CM, Hwu HG. Association between the dopamine transporter gene and the inattentive subtype of attention deficit hyperactivity disorder in Taiwan. Prog Neuropsychopharmacol Biol Psychiatry 2011;35:421ŌĆō428.


43. Correa-Noronha SA, Noronha SM, Alecrim C, Mesquita Ade C, Brito GS, Junqueira MG, et al. Association of angiotensin-converting enzyme I gene I/D polymorphism with endometrial but not with ovarian cancer. Gynecol Endocrinol 2012;28:889ŌĆō891.










