|
 |
- Search
| Genomics Inform > Volume 13(4); 2015 > Article |
Abstract
tRNA-derived RNA fragments (tRFs) are an emerging class of non-coding RNAs (ncRNAs). A growing number of reports have shown that tRFs are not random degradation products but are functional ncRNAs made of specific tRNA cleavage. They play regulatory roles in several biological contexts such as cancer, innate immunity, stress responses, and neurological disorders. In this review, we summarize the biogenesis and functions of tRFs.
It is now well known that mammalian genomes encode, in addition to protein-coding genes, RNA molecules that have no protein-coding capacity, and thus are referred to as non-coding RNAs (ncRNAs) [1]. Recent high throughput sequencing data suggest that there are about four times as many ncRNA transcripts as there are protein-coding RNA transcripts in human cells. While some ncRNAs, such as ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), and those well-studied ones including microRNAs (miRNAs) and small interfering RNAs (siRNAs), are recognized as functional molecules, the majority of ncRNAs and their biological roles in mammals are underappreciated, as evolution would have favored against such gratuitous and wasteful transcription [2]. However, accumulating evidence has exhibited a remarkable degree of their involvement in many biological events including cell proliferation, cellular stress responses, host immune responses and virus replication [3,4,5,6]. Based on their size, ncRNAs are classified into long, medium, and small ones. Those longer than 200 nucleotides (nt) are termed as long (or large) ncRNAs (lncRNAs) and include the majority of nuclear ncRNAs. Those ranging 40-200 nt are medium-sized ncRNAs including many classic ncRNAs such as 5S rRNA, tRNAs, small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), etc. ncRNAs shorter than 40 nt are termed small ncRNAs (sncRNAs) that have been extensively studied in the last two decades and will be elaborated in the next section. In this review, we will focus on a recently discovered class of sncRNAs called tRNA-derived RNA fragments (tRFs), as data from several groups including those from us suggested them to be emerging functional molecules.
The functionality of ncRNAs was initially bolstered by the discovery showing that sncRNAs are capable of mediating both transcriptional and post-transcriptional gene silencing [7]. The most extensively studied among them are miRNAs that regulate target mRNAs post-transcriptionally in animals and plants [8,9,10]. Another class of sncRNAs is siRNAs that are closely related to miRNAs [11]. Although naturally occurring siRNAs have been reported to be present in some mammals in addition to plants and flies [12,13], siRNAs are more renowned as an experimental tool in mammalian cells. The gene silencing by synthetic double-stranded RNAs (dsRNAs) and their Dicer-cleavage products (that are siRNAs) is called RNA interference (RNAi) and has been being used as a popular gene knockdown method [3]. This technical breakthrough was well recognized and celebrated by the 2006 Nobel Prize, which was awarded to the two scientists who discovered RNAi [14,15]. A number of other sncRNAs have also been identified in various organisms [16,17], including piwi-interacting RNA (piRNA), repeat-associated siRNA (rasiRNA), tiny ncRNAs (tncRNA), heterochromatic small RNA (21U-RNA, hcRNA), scan RNA (scnRNA), transcription initiation RNA (tiRNA) [18], and promoter/termini-associated sRNA (PASR/TASR) [19]. Although some of these have been shown to play roles in several biological processes such as heterochromatin formation and transposon silencing, their functions are largely unknown and detailed description of them is beyond the scope of this review [10].
Mature tRNAs are characterized by their evolutionarily conserved secondary and tertiary structures, extensive post-transcriptional modifications (up to 100 nucleoside modifications have been described), extreme stability and resistance to nucleases [20,21,22]. There are more than 600 tRNA genes in humans, which are far more than needed for at most 64 anticodons and 20 amino acids [23,24,25], suggesting gene-specific functions of tRNAs other than its traditional function in linking a codon to the cognate amino acid during translation. Indeed, tRFs were recently identified as a new class of sncRNAs fragmentized from tRNAs and shown to be important in regulating cell proliferation, stress-induced translational suppression, stress granule (SG) assembly, and virus replication. Expression of some tRFs is commonly seen among different organisms, further supporting that tRFs have emerged as important players in diverse aspects of biology.
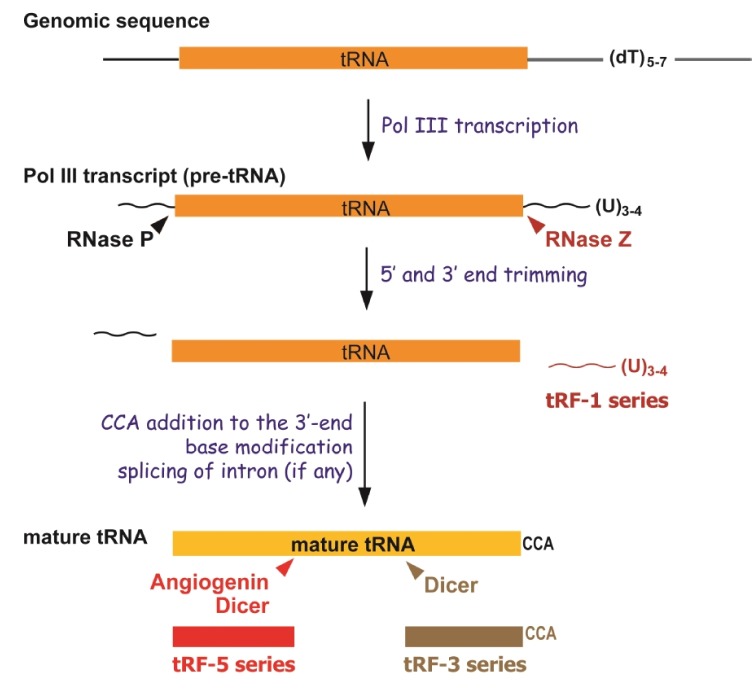
A mature tRNA has a 5'-monophosphate group and 3'-CCA charged with cognate amino acid. All properly folded tRNAs, with so called a cloverleaf structure, have four base-paired stems (in 5'–3' direction: D arm, anticodon arm, T arm, and acceptor stem) bridging the conserved D-loop, tRNA-specific anticodon loop, variable loop, and T-loop. Primary transcripts (pre-tRNAs) undergo several stages of maturation before becoming translation-competent tRNAs [26]. Compared to a mature tRNA, a pre-tRNA lacks a CCA tail but includes a 5'-leader starting with triphosphate, a 3'-trailer with an oligouridine stretch and, for intron-containing tRNA, a variably-sized intron. The 5'- and 3'-end of a pre-tRNA are trimmed by the endonucleases RNase P and RNase Z respectively, and an intron is spliced if there is [27,28]. The 'CCA' acceptor stem must be added to the 3'-end as this is not encoded in eukaryotes [29]. While mature, uncleaved tRNAs have a well-defined role in protein translation, multiple groups have cloned and sequenced shorter tRNA fragments [30]. These fragments (termed "tRFs") identified so far can be classified into three groups; tRF-5 series whose 5'-end is aligned to the 5'-end of mature tRNA, tRF-3 series whose 3'-end is aligned to the 3'-end of mature tRNA, and tRF-1 series which is the 3'-trailer sequence in a pre-tRNA and released during 3'-end processing (Fig. 1). Some groups have also termed the tRF-5 or tRF-3 series as tRF halves or type I [31,32] and tRF1 series as type II [32].
The tRF-5 series were the first identified class of tRFs and were usually formed by a cleavage in the D or anticodon loop of a mature tRNA. They have been reported in mammalian cells, plants, and fission yeast [3,33,34,35,36]. As aforementioned, the tRF-3 series are those derived from the 3'-end of a mature tRNA, usually by cleavage in the T loop [3,32,37,38,39]. As the 3'-end of mature tRNAs is the acceptor trinucleotide CCA, the tRF-3 series also characteristically has an end CCA. The tRF-1 series are undoubtedly the RNase Z cleavage product of a pre-tRNA, because their 5'-ends begin immediately after the 3'-terminus of the mature tRNA and their 3'-ends are a series of U residues that are residual from RNA polymerase III transcription run-off [3,32,40,41]. Overall, the cleavage of tRFs is very much structure-dependent, suggesting they are not randomly degraded products.
Other reasons supporting that tRFs are biologically meaningful include their endonuclease-specific cleavage [3,4,34,42]. The production of the tRF-5 series has been reported to be Dicer or angiogenin (ANG)-dependent in mammals [33,34]. In one case, the tRF-5 series were identified to be produced after the hybridization of the 3'-end of the human cellular tRNAlys with the HIV-1 primer binding site, raising the possibility that, at least in certain cases, tRFs may be produced when a tRNA base-pairs with another RNA molecule [38]. It has been reported that the tRF-3 series are generated by Dicer [32,38]. In Tetrahymena, although the nuclease identity was not proven, the tRF-3 series have 5'-monophosphates that are usually left after Dicer cleavage and are in physical association with Argonaute/Piwi proteins, suggesting Dicer-mediated cleavage [39]. The tRNA 3'-trailer sequences are thought to be rapidly degraded in the nuclear compartment soon after being cleaved from pre-tRNAs. Therefore, it is quite surprising that the tRF-1 series are localized mainly in the cytoplasm [3,41]. There are two explanations on how the tRF-1 series bypass nuclear cleavage: (1) the tRF-1 series are exported from the nucleus by an unknown mechanism upon processing, or (2) pre-tRNAs escape nuclear quality control and are processed by cytoplasmic RNase Z. Biological evidence indeed supports the second possibility. At least in one case of a tRF-1 molecule derived from tRNASer (termed tRF-1001 [3] and Cand45 [32]), the corresponding pre-tRNA is localized in the cytoplasm [3,32]. Also there are several reports supporting the cytoplasmic localization of tRNA endonucleases responsible for the tRF-1 series [3,43].
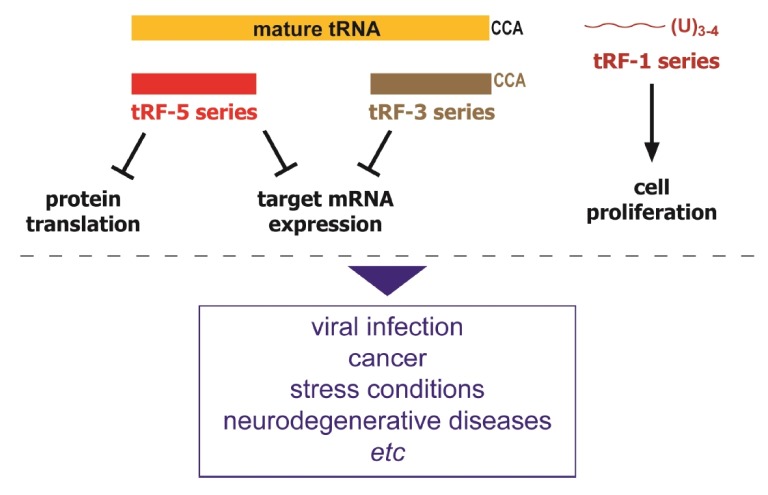
Are tRFs merely by-products of tRNA degradation or nuclease off-target effects? Or do they have bona fide biological functions? If they do have functions, how diverse are these functions? Several lines of evidence have identified their regulatory targets, supporting that they are functional molecules [31,34,35,37]. Although the exact roles of tRFs are further to be elucidated, accumulating evidence suggests that they participate in two main types of biological processes as discussed in more detail below (Fig. 2).
tRNAs are indispensable components of the translational machinery, hence one can surmise that tRNA cleavage affects protein synthesis. However, the mode of translational regulation by tRNA cleavage is not simply a result of change in the mature tRNA pool available for protein synthesis. Ivanov et al. showed that tRNA halves generated by ANG during stress play an active role in inhibiting protein synthesis and triggering the phospho-eIF2α-independent assembly of SGs [44].
In addition to tRNA halves, tRFs have also been reported to be involved in global translational inhibition in human cells. In the archaeon Haloferax volcanii a 26-nt long tRF-5 originating from tRNAVal in a stress-dependent manner was shown to directly bind to the small ribosomal subunit and reduce protein synthesis by interfering with peptidyl transferase activity [45]. A similar mechanism of translation inhibition by a tRF-5 was also observed in human cells [46]. A 19-nt long tRF-5 derived from tRNAGln decreased the expression of a reporter gene that did not have a sequence complementary to that of the tRF-5, suggesting that non-specific translational repression is mediated by this tRF-5. An interesting observation from this study was that the tRFs required a conserved "GG" dinucleotide for their activity in inhibiting translation. Because the tRF-5 series were detected in the polysome fraction, interaction of the tRF-5 with the ribosome may contribute to translation [46].
In addition to above-mentioned translational regulation, it has become apparent that some tRFs mediate gene silencing similarly to miRNAs by binding to Argonaute family proteins [16,17,47]. The first tRFs suggested to be involved in the gene silencing are some tRF-5 and -3 molecules [32]. Haussecker et al. observed that they are in association with Ago-3 and -4 and exhibit trans-gene silencing activity. However, this activity is much lower than that of miRNAs. On the other hand, some groups showed that the tRF-5 series have inefficient association with Argonautes [34] and do not exhibit detectable silencing effects [31].
Probably the most convincing result is from our recent studies on a tRF-5 molecule derived from mature tRNAGluCTC (tRF5-GluCTC). Upon infection of respiratory syncytial virus (RSV), tRF5-GluCTC is induced robustly and plays a gene-silencing role on a target mRNA with a reverse complementary target sequence. In this case, the trans-silencing mechanism used by this tRF was distinct from that of canonical miRNA/siRNA. Mutagenesis studies demonstrated that the 3'-portion of tRF5-GluCTC is critical for its gene silencing function, in contrast to miRNAs whose 5'-portion is the key determinant in target recognition [5].
The role of tRFs in cancer was first reported by Lee et al3. One of the tRF-1 series (namely tRF-1001), derived from pre-tRNASer, is highly expressed in several cancer cell lines relative to non-proliferating normal tissues, and is required for proliferation of prostate cancer cells [3]. The levels of tRF-1001 in cancer cells are directly related to the degree of cellular proliferation. Knocking down this tRF causes a dramatic loss of cell viability and inhibition of cell proliferation with specific accumulation of cells in G2 phase and inhibition of DNA synthesis [3]. In addition, analysis of the small RNA transcriptome in prostate cancer revealed enrichment of tRFs in both non-metastatic and metastatic lymph node prostate cancer samples [3]. The molecular mechanism by which tRF-1001 affects cell physiology is not known. There may be a differential processing of tRNA in prostate cancer because tRFs in non-metastatic samples were 18 nucleotides long whereas tRFs in metastatic tissues were 27 nucleotides long [49].
The important role of tRFs in controlling cancer is also supported by the observation showing enhanced ANG expression in almost all types of cancer [50,51,52]. Recently, it has been shown that ANG produced tRFs that re-program translation and promote the assembly of stress granules. As these events help cells survive under adverse conditions, it is possible that ANG-induced tRFs may contribute to ANG-mediated cancer cell proliferation [48]. tRFs have also been reported to help cancer cells to prevent apoptosome formation by binding to cytochrome c [53].
Although tRNA metabolism is essential for all human cells, a number of neurological disorders are caused by defects in tRNA metabolism and tRNA processing enzymes. ANG mutants possessing reduced RNase activity were implicated in the pathogenesis of amyotrophic lateral sclerosis (ALS), a fatal neurodegenerative disease [54]. A subset of ALS-associated ANG mutants was also found in Parkinson's disease patients [55]. Administration of recombinant ANG to cultured neurons protects them from stress-induced apoptosis and promotes the lifespan and motor function of SOD1 (G93A) mice, a laboratory ALS model.
In neurons, excessive accumulation of some tRF-5 series members that are derived from a specific subset of tRNAs (Asp, Glu, Gly, His, Val, and Lys) triggers a sustained stress response leading to neuronal loss and links aberrant tRNA metabolism to the development of certain forms of intellectual disability [48].
tRFs derived from intron-containing tRNAs are also strongly implicated in neurodegeneration [56,57,58]. A study on mutant mice carrying a catalytically inactive form of the RNA kinase CLP1, a component of the tRNA splicing endonuclease complex, revealed a neurological disorder characterized by progressive loss of lower spinal motor neurons. Surprisingly, mutant mice accumulate a novel class of tRFs. In addition, homozygous missense mutations in CLP1 (R140H) were found in patients with pontocerebellar hypoplasia, a heterogeneous group of inherited neurodegenerative disorders characterized by impaired development of various parts of the brain. These patients were identified to suffer from severe motor-sensory defects, cortical dysgenesis and microcephaly, and exhibit alterations in tRNA splicing [59]. On the cellular level, the R140A mutation leads to the depletion of mature tRNAs, the accumulation of unspliced pre-tRNAs in patient-derived induced neurons [58] and the accumulation of linear tRNA introns [57]. Transfection of 5'-unphosphorylated tRF corresponding to the 3'-exon of pre-tRNATyr (the natural substrate of CLP1) into patient cells results in reduced neuron survival under oxidative stress compared to the transfection of the 5'-exon, which had no effect on viability [58]. The kinase-dead CLP1 mice accumulate 5'-leader-exon-tRFs that sensitize cells to oxidative stress and promote cell death in a p53-dependent fashion [56]. While the exact mechanistic details on the interplay between CLP1 activity, tRNA splicing and functions of pre-tRNA-derived tRFs are yet to be uncovered, these studies provide a basis for further investigations linking aberrant tRNA metabolism and development of neurodegeneration [48].
Viruses exploit a number of strategies to evade host immune recognition and to facilitate viral replication. Some of these strategies involve viral-derived or endogenous cellular ncRNAs that interact with host immune responses [60].
We have demonstrated that RSV, the most common cause of bronchiolitis and pneumonia in babies, employs a novel mechanism to suppress antiviral responses via induction of some tRFs [5,61]. We have shown that RSV infection in airway epithelial cells leads to the activation of endonuclease ANG, subsequently inducing abundant production of tRFs, most of which belong to that tRF-5 series. The cleavage sites by ANG are preferentially adjacent to the 5'-end of the anticodon-loop, generating tRF-5 series. These tRF-5s are localized exclusively in the cellular cytoplasm [5]. At least one of the tRFs, namely tRF5-GluCTC (tRF-5 derived from tRNAGlu), has a trans-gene silencing function. Significantly, an antiviral target suppressed by tRF5-GluCTC downregulates RSV replication and significantly decreases the yield of infectious viral particles produced by infected cells, demonstrating a stimulatory role for tRF5-GluCTC in RSV replication [5].
Using an elegant biochemical approach, named HITS-ABt (hIgh-throughput sequencing of RNAs associated with biotinylated tRF), to capture tRF5-GluCTC-containing ribonucleoprotein complexes combined with bioinformatics analysis, we have identified a number of potential cellular mRNA targets of tRF5-GluCTC. One of the candidate mRNAs was shown to be a direct target of tRF5-GluCTC. Mechanistically, tRF5-GluCTC recognizes a target site in the 3'-untranslated region of APOER2 mRNA and suppresses its expression by an unknown mechanism. Further analysis identified a previously unappreciated role for APOER2 protein in antiviral defense against RSV infection [61]. Thus, we have illuminated the roles of tRFs in host–virus interactions and described a novel molecular mechanism for host response regulation via gene targeting at posttranscriptional level, proving a potential therapeutic target to control RSV replication by regulating tRFs induction. As another example of tRFs with a positive effect on viral replication, a tRF-3 derived from tRNAPro (tRF-3019) has been suggested to function as a primer for reverse transcription of human T-cell leukemia virus-1 [62].
The induction of tRFs is virus specific, as human metapneumovirus, a negative-sense single stranded RNA virus belonging to the Paramyxoviridae family [63] and frequent cause for lower respiratory tract infections in young children, immunocompromised patients and older adults [64,65,66,67], did not induce tRFs but gradually enhanced the expression percentage of miRNAs in a time-dependent fashion [6]. The involvement of tRFs in infectious diseases suggests that tRFs may be targets for the development of new drugs to control viral replication.
The human genome encodes hundreds of tRNA genes and pseudogenes. In the past two decades, an increasing number of reports have revealed abundant expression of functional tRFs; thus, it now appears highly plausible that cells use tRNAs as a source for functional sncRNAs to modulate biological processes beyond its translation function. It is imperative to discriminate functional tRNA fragments from "meaningless" degradation products, and to biochemically elucidate the factors in their biogenesis as well as to determine their molecular functions. Although tRFs, at least the tRF-5s induced by RSV, regulate gene expression at post-transcriptional stages in the cytoplasm, the precise mechanism is yet to be clarified. Like miRNAs, tRFs can repress target mRNAs by inhibiting their translation or facilitating their decay. These questions can be answered by analyzing their localization, identifying their interacting protein partners besides Argonaute proteins, and examining various biological phenomena in response to the changes in the expression of tRFs. Bioinformatics studies with the support from biochemical assays have identified numerous novel tRNA-interacting proteins [68], implying that the biological functions of tRNAs and their fragments may be way beyond our expectations. Further combination of computational and biochemical efforts will significantly advance our understanding of functional tRFs and expand our knowledge regarding the tRNA world.
The availability of genome sequencing data from patients with diverse conditions has already led to the identification of several mutations in tRNA genes and in genes involved in tRNA biogenesis [69]. The task now is to ascertain whether these mutations indeed affect the repertoire of tRFs and consequently the molecular pathology of the diseases. Studies on tRFs are still at a very early stage. Additional functions for tRFs are likely to be discovered continuously. Improved understanding of the functions of tRFs will provide valuable insights into human physiology and pathophysiology.
Recognition of tRFs as an important class of functional molecules has not happened until recently. The apparent presence of abundant tRFs in early studies with next generation sequencing data was often ignored. However, the combined biochemical evidence from many years of tRNA biology studies has brought the field to a previously unexpected conceptual consensus: specific tRFs are not simply random degradation products but functional molecules based on the following observations: (1) in many cases, tRFs do not derive from abundant cellular tRNAs, and the numbers of tRFs do not correlate with the parental tRNA gene copy numbers; (2) their fragmentation patterns are tRNA structure-dependent; (3) the fragmentation patterns is determined by the developmental stage or cell conditions; and (4) it is reported that some tRFs are bound to Argonaute/Piwi proteins, well-known components of the RNA-induced silencing complex [21,31,70,71,72,73]. The expression of tRFs does not usually affect mature tRNA pools. Instead, it is involved in normal biological processes beyond translation and in diseases; thus, studies of tRFs have provided new insights into tRNA biology.
References
1. Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nat Rev Genet 2010;11:559–571. PMID: 20628352.


2. Ponting CP. The functional repertoires of metazoan genomes. Nat Rev Genet 2008;9:689–698. PMID: 18663365.


3. Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 2009;23:2639–2649. PMID: 19933153.



4. Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 2010;285:10959–10968. PMID: 20129916.



5. Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther 2013;21:368–379. PMID: 23183536.



6. Deng J, Ptashkin RN, Wang Q, Liu G, Zhang G, Lee I, et al. Human metapneumovirus infection induces significant changes in small noncoding RNA expression in airway epithelial cells. Mol Ther Nucleic Acids 2014;3:e163. PMID: 24845106.



7. Morris KV. RNA-directed transcriptional gene silencing and activation in human cells. Oligonucleotides 2009;19:299–306. PMID: 19943804.



8. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–297. PMID: 14744438.


9. Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development 2005;132:4645–4652. PMID: 16224044.


11. Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001;411:494–498. PMID: 11373684.


12. Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet 2007;8:884–896. PMID: 17943195.


13. Nilsen TW. Endo-siRNAs: yet another layer of complexity in RNA silencing. Nat Struct Mol Biol 2008;15:546–548. PMID: 18523465.


15. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998;391:806–811. PMID: 9486653.


16. Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet 2009;10:94–108. PMID: 19148191.



17. Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 2008;135:1201–1214. PMID: 18287206.


18. Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet 2009;41:572–578. PMID: 19377478.


19. Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007;316:1484–1488. PMID: 17510325.


20. Giege R. Toward a more complete view of tRNA biology. Nat Struct Mol Biol 2008;15:1007–1014. PMID: 18836497.


21. Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev 2010;24:1832–1860. PMID: 20810645.



22. Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol Med 2014;20:306–314. PMID: 24581449.


23. Abe T, Inokuchi H, Yamada Y, Muto A, Iwasaki Y, Ikemura T. tRNADB-CE: tRNA gene database well-timed in the era of big sequence data. Front Genet 2014;5:114. PMID: 24822057.



24. Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 2009;37:D93–D97. PMID: 18984615.


25. Parisien M, Wang X, Pan T. Diversity of human tRNA genes from the 1000-genomes project. RNA Biol 2013;10:1853–1867. PMID: 24448271.



26. Li H. Complexes of tRNA and maturation enzymes: shaping up for translation. Curr Opin Struct Biol 2007;17:293–301. PMID: 17580114.


27. Mitchell HD, Eisfeld AJ, Sims AC, McDermott JE, Matzke MM, Webb-Robertson BJ, et al. A network integration approach to predict conserved regulators related to pathogenicity of influenza and SARS-CoV respiratory viruses. PLoS One 2013;8:e69374. PMID: 23935999.



28. Schiffer S, Rosch S, Marchfelder A. Assigning a function to a conserved group of proteins: the tRNA 3'-processing enzymes. EMBO J 2002;21:2769–2777. PMID: 12032089.



29. Aebi M, Kirchner G, Chen JY, Vijayraghavan U, Jacobson A, Martin NC, et al. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J Biol Chem 1990;265:16216–16220. PMID: 2204621.


31. Sobala A, Hutvagner G. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip Rev RNA 2011;2:853–862. PMID: 21976287.


32. Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010;16:673–695. PMID: 20181738.



33. Buhler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol 2008;15:1015–1023. PMID: 18776903.



34. Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009;15:2147–2160. PMID: 19850906.



35. Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 2009;151:2120–2132. PMID: 19854858.



36. Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Suzuki H, Hayashizaki Y, et al. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol 2011;8:158–177. PMID: 21282978.



37. Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, et al. Hidden layers of human small RNAs. BMC Genomics 2008;9:157. PMID: 18402656.



38. Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res 2009;37:6575–6586. PMID: 19729508.



39. Couvillion MT, Sachidanandam R, Collins K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev 2010;24:2742–2747. PMID: 21106669.



40. Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 2008;22:2773–2785. PMID: 18923076.



41. Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, et al. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3' trailers. PLoS One 2010;5:e10563. PMID: 20498841.



42. Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 2009;185:35–42. PMID: 19332886.



43. Elbarbary RA, Takaku H, Uchiumi N, Tamiya H, Abe M, Nishida H, et al. Human cytosolic tRNase ZL can down-regulate gene expression through miRNA. FEBS Lett 2009;583:3241–3246. PMID: 19751732.


44. Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 2011;43:613–623. PMID: 21855800.



45. Gebetsberger J, Zywicki M, Kunzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012;2012:260909. PMID: 23326205.




46. Sobala A, Hutvagner G. Small RNAs derived from the 5' end of tRNA can inhibit protein translation in human cells. RNA Biol 2013;10:553–563. PMID: 23563448.



47. Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009;10:126–139. PMID: 19165215.


48. Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett 2014;588:4297–4304. PMID: 25220675.



49. Martens-Uzunova ES, Jalava SE, Dits NF, van Leenders GJ, Moller S, Trapman J, et al. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene 2012;31:978–991. PMID: 21765474.


50. Tello-Montoliu A, Patel JV, Lip GY. Angiogenin: a review of the pathophysiology and potential clinical applications. J Thromb Haemost 2006;4:1864–1874. PMID: 16961595.


51. Gao X, Xu Z. Mechanisms of action of angiogenin. Acta Biochim Biophys Sin (Shanghai) 2008;40:619–624. PMID: 18604453.


52. Li S, Hu GF. Emerging role of angiogenin in stress response and cell survival under adverse conditions. J Cell Physiol 2012;227:2822–2826. PMID: 22021078.



53. Saikia M, Jobava R, Parisien M, Putnam A, Krokowski D, Gao XH, et al. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol 2014;34:2450–2463. PMID: 24752898.



54. Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, et al. ANG mutations segregate with familial and 'sporadic' amyotrophic lateral sclerosis. Nat Genet 2006;38:411–413. PMID: 16501576.


55. van Es MA, Schelhaas HJ, van Vught PW, Ticozzi N, Andersen PM, Groen EJ, et al. Angiogenin variants in Parkinson disease and amyotrophic lateral sclerosis. Ann Neurol 2011;70:964–973. PMID: 22190368.



56. Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature 2013;495:474–480. PMID: 23474986.



57. Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, et al. Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell 2014;157:636–650. PMID: 24766809.



58. Schaffer AE, Eggens VR, Caglayan AO, Reuter MS, Scott E, Coufal NG, et al. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 2014;157:651–663. PMID: 24766810.



59. Weitzer S, Hanada T, Penninger JM, Martinez J. CLP1 as a novel player in linking tRNA splicing to neurodegenerative disorders. Wiley Interdiscip Rev RNA 2015;6:47–63. PMID: 25142875.


60. Li F, Ding SW. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol 2006;60:503–531. PMID: 16768647.



61. Deng J, Ptashkin RN, Chen Y, Cheng Z, Liu G, Phan T, et al. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol Ther 2015;23:1622–1629. PMID: 26156244.



62. Ruggero K, Guffanti A, Corradin A, Sharma VK, De Bellis G, Corti G, et al. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. J Virol 2014;88:3612–3622. PMID: 24403582.



63. van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 2001;7:719–724. PMID: 11385510.



64. Edwards KM, Zhu Y, Griffin MR, Weinberg GA, Hall CB, Szilagyi PG, et al. Burden of human metapneumovirus infection in young children. N Engl J Med 2013;368:633–643. PMID: 23406028.



65. Englund JA, Boeckh M, Kuypers J, Nichols WG, Hackman RC, Morrow RA, et al. Brief communication: fatal human metapneumovirus infection in stem-cell transplant recipients. Ann Intern Med 2006;144:344–349. PMID: 16520475.


66. Esper F, Martinello RA, Boucher D, Weibel C, Ferguson D, Landry ML, et al. A 1-year experience with human metapneumovirus in children aged <5 years. J Infect Dis 2004;189:1388–1396. PMID: 15073675.


67. Falsey AR, Erdman D, Anderson LJ, Walsh EE. Human metapneumovirus infections in young and elderly adults. J Infect Dis 2003;187:785–790. PMID: 12599052.


68. Parisien M, Wang X, Perdrizet G 2nd, Lamphear C, Fierke CA, Maheshwari KC, et al. Discovering RNA-protein interactome by using chemical context profiling of the RNA-protein interface. Cell Rep 2013;3:1703–1713. PMID: 23665222.



69. Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet 2015;16:98–112. PMID: 25534324.


70. Raina M, Ibba M. tRNAs as regulators of biological processes. Front Genet 2014;5:171. PMID: 24966867.



71. Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol 2014;12:78. PMID: 25270025.



72. Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol 2013;10:1798–1806. PMID: 24351723.



73. Garcia-Silva MR, Cabrera-Cabrera F, Guida MC, Cayota A. Hints of tRNA-Derived Small RNAs Role in RNA Silencing Mechanisms. Genes (Basel) 2012;3:603–614. PMID: 24705078.



- TOOLS
-
METRICS

-
- 42 Crossref
- 9,057 View
- 148 Download
- Related articles in GNI