 |
 |
- Search
| Genomics Inform > Volume 12(1); 2014 > Article |
Abstract
Asian populations contain a variety of ethnic groups that have ethnically specific genetic differences. Ethnic variants may be highly relevant in disease and human differentiation studies. Here, we identified ethnically specific variants and then investigated their distribution across Asian ethnic groups. We obtained 58,960 Pan-Asian single nucleotide polymorphisms of 1,953 individuals from 72 ethnic groups of 11 Asian countries. We selected 9,306 ethnic variant single nucleotide polymorphisms (ESNPs) and 5,167 ethnic variant copy number polymorphisms (ECNPs) using the nearest shrunken centroid method. We analyzed ESNPs and ECNPs in 3 hierarchical levels: superpopulation, subpopulation, and ethnic population. We also identified ESNP- and ECNP-related genes and their features. This study represents the first attempt to identify Asian ESNP and ECNP markers, which can be used to identify genetic differences and predict disease susceptibility and drug effectiveness in Asian ethnic populations.
There has been an explosion of data describing genetic variants in humans. Structural genetic variations, such as single nucleotide polymorphisms (SNPs) and copy number variations (CNVs), have given rise to myriad differences in human populations [1]. The study of human genetic variations has both evolutionary significance and medical applications and can help scientists understand ancient human population migrations as well as how different human groups are biologically related to one another [2, 3]. SNPs represent the most frequent type of human DNA variation [4]. The main goals of SNP research include understanding the genetics of the human phenotype variation and, especially, the genetic basis of human diseases [5, 6]. Genome-wide linkage and association studies have been made possible by highly accurate methods for typing SNPs [7]. CNV is a form of genomic structural variation that results in the cell having an abnormal number of copies of one or more sections of DNA [8]. CNVs can be limited to a single gene or include a contiguous set of genes. CNVs can result in having either too many or too few dosage-sensitive genes, which may be responsible for a substantial amount of human phenotypic variability, complex behavioral, traits and disease susceptibility [9]. A copy number polymorphism (CNP) is a CNV that occurs in more than 1% of the population [10]. SNP platforms can also be used for typing CNVs. This allows for generalized genotyping of both SNPs and CNVs simultaneously on a common sample set, with advantages in terms of cost and unified analysis. Although CNP detection from SNP genotyping data is a difficult task and has the limitation of false positive results, it will provide a more comprehensive view of genomic variations and complement genome-wide association studies in identifying disease susceptibility loci.
SNPs are being used in studies of human migration and evolution, as well as those of human health. The Human Genome Organization (HUGO) Pan-Asian SNP Consortium reported a large-scale survey of autosomal variations from a broad geographic sample of 72 Asian human populations [11]. The study indicated that most populations show relatedness within ethnic or linguistic groups, despite significant gene flow among groups. This relatedness may have important implications for our understanding of genetics and disease. Data on ethnic populations in the Pan-Asia region can be valuable to show the spectrum of genetic diversity and networks of ethnic groups. Thus, notwithstanding the population size problem in some ethnic groups, we investigated the ethnic specificity based on SNP and CNP information. Here, we identified genetic variations-ethnically specific SNPs (ESNPs) and ethnically specific CNPs (ECNPs)-of Asian populations using SNP genotypic profiling. These ESNP and ECNP markers can be used to identify genetic differences and to predict disease susceptibility and drug effectiveness in Asian populations.
We obtained a genome-wide 58,960-SNP dataset (Affymetrix GeneChip Human Mapping 50K Xba chip; Affymetrix Inc., Santa Clara, CA, USA) from the HUGO Pan-Asian SNP Consortium website (http://www4a.biotec.or.th/PASNP) [12]. The SNP data were obtained from 1,833 individual DNA samples collected from blood and buccal samples of 72 Asian and non-Asian ethnic groups. Filtering SNPs is an important step in genome-wide studies to minimize potential false findings. We filtered SNPs at 2 levels: individual and SNP. In the individual-level filtering step, we removed individual SNPs that had a <90% genotyping call rate and that were not in Hardy-Weinberg equilibrium (p < 0.001). We filtered out 89 of the initial 1,833 individuals and used SNPs from the 1,744 individuals for further study. In the SNP-level filtering step, 1,692 of the initial 58,960 SNPs were filtered out, because 389 SNPs had an SNP call rate below 90%, 451 SNPs were physically unmapped, 103 SNPs could not be mapped to dbSNP, and 749 SNPs were monomorphic. Finally, we obtained 56,025 SNP markers from 1,744 genotyped individuals, representing 72 ethnic groups in the HUGO Pan-Asian SNP Consortium.
For comparison with other ethnic variants, we obtained SNPs of 209 HapMap individuals representing 4 populations (China Han [CHB]; Japan Japanese [JYP]; USA European [CEU]; Africa Yoruba [YRI]) (http://www.hapmap.org/). The 243 SNPs in the HUGO Pan-Asian with the HapMap dataset were nonshared SNP loci and excluded.
Phasing is needed to determine which variants are inherited by an individual at each locus and to more accurately determine the relationships between unrelated individuals in large population datasets. Using phasing population data, we can identify haplotypes, which are essentially segments of DNA that are common to a particular ethnic group. The fastPHASE program version 1.1.4 [13] was used to estimate missing genotypes and reconstruct haplotypes from unphased SNP genotype data of unrelated individuals in order to identify ethnicity-specific SNPs.
SNP genotyping arrays recently have been used for CNP detection and analysis, because the arrays can serve dual roles for SNP- and CNV-based association studies. To detect CNP markers from the SNPs, we used the Affymetrix Genotyping Analysis Software and Copy Number Analysis Tool (CNAT version 3.0), which was downloaded from the Affymetrix website (http://www.affymetrix.com/products/software). We calculated the distribution of genomic smoothing copy number signal intensity (upper-boundary = 2.78, lower-boundary = 1.51) to detect the CNPs (Supplementary Fig. 1). We regarded markers with values beyond the upper- and lower-boundary values as CNVs. Then, we selected the CNPs that presented variation in >1% of the individuals as CNVs that occurred in more than 1% of the population [14]. We downloaded the detection tools from the website (http://www.affymetrix.com/products/software) and used these programs for our Pan-Asian SNP data. We regarded markers with values outside of the upper- and lower-boundary values as CNVs. Then, we selected CNPs which presented variation in >1% of the 1,196 individuals.
To investigate the distribution of ESNP and ECNP markers in Pan-Asian ethnic groups, we performed the following steps, as shown in Supplementary Fig. 2. First, we classified the ethnic groups into 3 hierarchical categories by population estimation of the phylogenic analysis (in Supplementary Fig. 2): superpopulation (clustering size = 4), subpopulation (ESNP clustering size = 12, ECNP clustering size = 11), and ethnic population (ESNP clustering size = 72, ECNP clustering size = 60). The clustering size in parentheses refers to how many distinct populations were used in the ESNP and ECNP analysis. The superpopulations consisted of African, Caucasian, American Indian, Asian, and outlier (Indian and Uyghur) populations. The subpopulations comprised population groups based on linguistic similarities and population structures and 2 additional groups (Mongoloid features [IN-NI]/Mongoloid features [IN-TB] and China Uyghur [CN-UG]), and the ethnic populations represented each ethnic group. Second, we identified ethnicity-specific variations based on SNPs and CNPs markers using the nearest shrunken centroid method (NSCM), which is not affected by the minor-allele frequency [15, 16]. The NSCM of the R package pamr carried out multiple tests for genetic heterogeneity and frequency spectrum on genes having ethnicity-specific variants. It shrinks each of the class centroids toward the overall centroid for all classes by a threshold. This shrinkage makes the classifier more accurate by eliminating the effect of noise. This method is a suitable attempt to solve the classification problem when there are a large number of features from which classes and a relatively small number of cases are predicted, and it is a significant approach to identify which features contribute most to the classification [15]. Third, we identified specific ethnic groups that correlated with each SNP or CNP. Finally, to investigate the characteristics of ethnicity-specific variations, we obtained the top 10% of ESNPs and ECNPs markers.
To investigate the characteristics of the ECNP- and ESNP-related genes, we selected the ECNP and ESNP markers ranked in the top 1% of the ethnically specific variations. We mapped ESNP and ECNP markers to gene structure, based on Entrez gene-centered information at NCBI [16]. We identified the relationships among ESNP- and ECNP-genes using the Ingenuity Systems Pathway Analysis tool (http://www.ingenuity.com), which is a web-based tool for pathway and network analysis of genes.
We obtained 58,960 Pan-Asian SNPs from 72 Pan-Asian populations listed in the supplementary documents (Supplementary Table 1). The SNP dataset shows the spectrum of genetic and phenotypic diversity in Asian populations and allows us determine the ethnic specificity of Asian populations based on ESNP and CNP makers.
We devised an approach to identify ethnic differences from the genotype profiles of the populations. Pan-Asian SNPs were filtered using several steps, as described in Materials and Methods. With the filtered SNPs, we examined the inter-SNP distances, the allele frequency, and the heterozygosity distribution to identify the genotypic features of each ethnic group. The average inter-SNP distance was 52 kb. More than 41% of SNPs fell below 10 kb, and 14% was over 100 kb (Supplementary Fig. 3). The inter-SNP distance distribution within 10 kb of one another was similar to the distribution observed in the HapMap data.
We investigated minor allele frequency and heterozygosity across the ethnic groups (Supplementary Fig. 4). We compared SNP genotypes in the ethnic groups with those identified by the HapMap [17] consortium. The proportion of Pan-Asian SNPs in each minor allele frequency and heterozygosity range was similar to those of the 4 HapMap ethnic groups. In particular, the SNP proportions for CN-UG, Proto-Austroloids (IN-DR), and Singapore Indian (SG-ID) were almost the same as those of the 4 HapMap ethnic groups. We found that 5 ethnic groups (America Melanesians [AX-ME], Thailand Mlabri [TH-MA], Indonesia Mentawai [ID-MT], CN-UG, and SG-ID) had minor allele frequency and heterozygosity distributions that were significantly different from those of other ethnic groups. Approximately 40% of the SNPs in the AX-ME, TH-MA, and ID-MT populations fell into the <0.05 range. The proportion of SNPs with heterozygosity <0.05 was lowest in the CN-UG and SG-ID groups, whereas the proportion of SNPs with heterozygosity <0.10 was highest in the TH-MA and AX-ME groups.
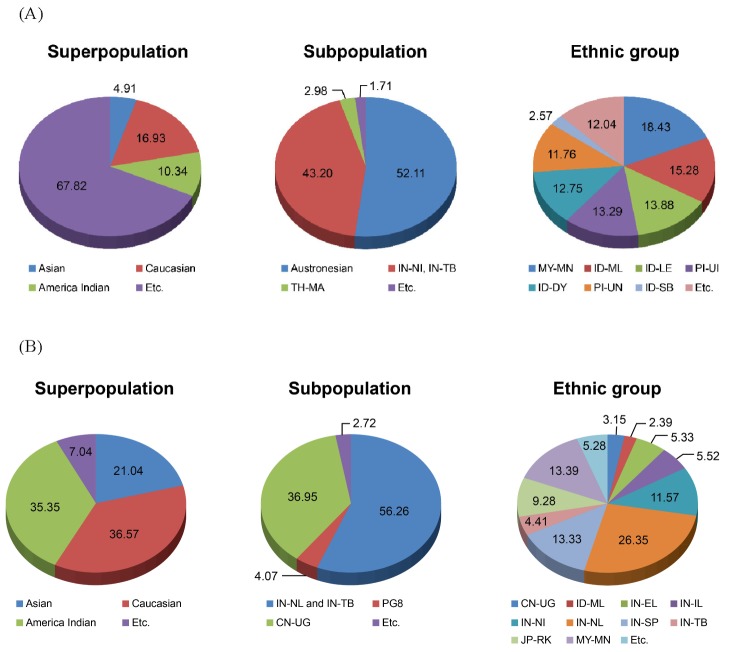
We confirmed the high proportion of African (YRI)-specific SNPs from 3 races: Asian, Caucasian, and African (Supplementary Fig. 5). Then, to search ESNPs from Pan-Asian ethnic groups, we identified 9,306 ESNPs from the Pan-Asian SNP markers and examined the distribution of the ESNPs at the 3 population levels: super-, sub-, and ethnic population (Fig. 1A). We found that 67.82% of ESNPs in the superpopulation level were others, including IN-NI, IN-TB, and CN-UG. It could show that isolated ethnic groups keep their original features and that these features are also presented in other population levels. In the subpopulations, Austronesian, IN-NI, and IN-TB had 95.31% of ESNPs. Additionally, we identified 28,282 CNPs based on the SNP chip profiles and then selected 5,167 ECNPs. We then examined the ECNPs in the 3 population levels (Fig. 1B). In the superpopulations, 36.57% of the ECNPs occurred in Caucasoids. Of the subpopulation ECNPs, 56.26% had Mongoloid features and 36.95% occurred in Uyghurs. Across the ethnic groups, 26.35% of the ECNPs were from Caucasoids (IN-NL). Most ECNPs occurred in Caucasoid populations (IN-NL, IN-IL, IN-DR, IN-SP, IN-EL, IN-WI, IN-WL, and SG-ID) and in ethnic groups that have Mongoloid features (IN-NI, IN-TB, and CN-UG). The ethnic group having Mongoloid features had more ECNPs than other groups, which shows that Pan-Asian ECNPs could serve as the criteria for ethnic specificity in Pan-Asian populations.
We obtained genes associated with ESNPs and ECNPs based on the NCBI Gene database [18]. We identified 156 ESNP-related genes and 52 ECNP-related genes (Supplementary Table 2). Most of the ESNP-containing genes were associated with known molecular functions encoded for cellular assembly and maintenance, cellular movement, cell death and survival, and lipid metabolism (Table 1). These ESNP-related gene sets (CEP192, GRK5, TUBGCP3, TUBGCP6, ABCA1, CHGA, ITGB3, VAV2, MYLK3, ANK2, ITGB3, ELN, LTBP4, FAM107B, DIDO1, HOXD3, PRPX2, KCNMA1, RASGRF1, NF1, and UBR2) showed association with diseases, such as cardiovascular disease, dermatological diseases and conditions, developmental disorders, and connective tissue disorders. Defects in latent transforming growth factor beta binding protein 4 (LTBP4) may be a cause of cutis laxa and severe pulmonary, gastrointestinal, and urinary abnormalities [19]. Centrosomal protein 192 kDa (CEP192) is part of a large multisubunit complex required for microtubule nucleation at the centrosome. This gene is regulated by hepatitis B virus and has roles in cells, such as duplication, replication, assembly, and nucleation. The ECNP-related genes are associated with known molecular functions encoded for cell-to-cell signaling and interaction, molecular transport, and drug metabolism (Table 1). These ECNP-containing genes (ARID1B, CAPN7, DDR2, SLC22A1, SLC22A3, ASTN2, GABRA5, GABRG3, RBFOX1, TLR4, MAG, TLR4, SNX9, and SYNJ2) are associated with cardiovascular disease, neurological disease, psychological disorders, and developmental disorders. Interestingly, the APOBEC gene, encoding the CŌåÆU editing enzyme family, such as apolipoprotein B mRNA editing enzyme and catalytic polypeptide-like 2, is associated with ECNPs. This editing gene could be affected by genetic diversity and admixtures in Pan-Asian populations. In this ethnically specific genetic variation study, we found the features of gene sets having ethnically specific variations. The AT-rich interactive domain 1B (SWI1-like) (ARID1B) gene encodes an AT-rich DNA interaction domain-containing protein. This protein is a component of the SWI/SNF chromatin remodeling complex and may play a role in cell-cycle activation. It is associated with chromatin-mediated maintenance of transcription and nervous system development. This gene is related to autosomal dominant mental retardation type 12. Discoidin domain receptor tyrosine kinase 2 (DDR2) is associated with colorectal neoplasm, metastatic colorectal cancer, hepatocellular carcinoma, gastrointestinal stromal tumor, and so on. These ethnically specific genetic variations represent relationships with cell cycling, metabolism, and the developmental and immune systems. These genetic variations can be affected by admixture and evolution. These ethnically specific variants may be forced to adapt in order to survive in new environments and play a significant role in development and functional systems associated with diseases [20].
Our analysis was able to identify ethnically variable SNPs associated with phenotypic changes. We selected 9,306 ESNPs and 5,167 ECNPs in 72 Pan-Asian populations. We found that representative ethnic groups with specific ESNPs are recently branched-out subpopulations, whereas representative ethnic groups with ethnic specific CNPs are early fixed subpopulations, as shown in Supplementary Fig. 6. This likely occurred due to accelerated and accumulated genetic drifts or selective pressure. This shows that ESNPs may participate in the ongoing creation of genetic variation through selective pressure or selective sweep. These ethnically specific variations are associated with 156 ESNP-related genes and 52 ECNP-related genes. Ethnically specific genetic variations may affect phenotype variation and disease susceptibility. Although it is not enough information to compare ECNP distribution and ESNP distribution directly, significant gene sets having ESNPs or ECNPs can be useful to study disease risk and drug susceptibility.
Acknowledgments
This research was supported by a grant from the KRIBB Research Initiative Program and by the Korean Ministry of Science, ICT & Future Planning (MSIP) under grant number 2013036118 (NRF-2011-0019745). Authors thank Ms. Kyeyoung Kim for editing the figures.
Supplementary materials
Supplementary data including two tables and six figures can be found with this article online at http://www.genominfo.org/src/sm/gni-12-42-s001.pdf.
Supplementary Information
Identification of Ethnically Specific Genetic Variations in Pan-Asian Ethnos
Supplementary Table 1
Ethnic groups list in Pan Asia (assign Ethnic group as like "two country-two ethnic group") and HapMap populations
Supplementary Fig. 1
Distribution of Pan-Asian copy number values from Pan-Asian genotype profiling. It shows that discrete distribution of Pan-Asian single nucleotide polymorphism samples with lower boundary of 1.5065 and upper boundary of 2.7765.
Supplementary Fig. 2
Process to select ethnic specific single nucleotide polymorphism (SNP). NSCM, nearest shrunken centroid method.
Supplementary Fig. 3
Inter-single nucleotide polymorphism (SNP) distance distribution. The X-axis represents the distance (kilo base pair) between SNPs and the Y-axis represents the proportion (%) of pan Asian SNPs.
Supplementary Fig. 4
Minor allele frequency (MAF) and heterozygosity (HET) distribution of single nucleotide polymorphism (SNP) in the Pan-Asian SNP data. X-axis is Pan-Asian ethnic groups, and Yaxis is SNP proportion (%) for MAF in each range. We examined MAF and HET across ethnic groups and 4 HapMap groups together. We assigned SNP proportion (%) in each range for MAF and HET rate of Pan-Asia ethnic groups and 4 HapMap groups as shown in (A) and (B).
Supplementary Fig. 5
The distribution of ethnic variant single nucleotide polymorphisms (ESNPs) across Pan-Asian and HapMap ethnic groups. We analyzed the ethnicity-specific single-nucleotide polymorphisms, including the four HapMap groups (CEU, CHB, JPT, and YRI), for the following groups: super-populations (Asians, Caucasoids, American Indians, and outliers [IN-NI, IN-TB, and CN-UG]); 12 populations; and 76 ethnic groups (Pan-Asian and four HapMap ethnic groups). AX-AI, Karitiana; AX-ME, Ami; CEU, European; PI-AE, Ayta; PI-AG, Ayta; PI-MW, Mamanwa; TH-MA, Mlabri; YRI, Yoruba; CHB, Han; JPT, Japanese; IN-NI, Mongoloid features; IN-TB, Mongoloid features; CN-UG, Uyghur.
Supplementary Fig. 6
Representative ethnic groups having ethnically specific single nucleotide polymorphismss and copy number polymorphisms on population structures. Yellow-colored row indicates the Pan-Asian ethnic groups having highly portion of ethnic variant single nucleotide polymorphisms and red-colored row indicates the Pan-Asian ethnic groups having highly portion of ethnic variant copy number polymorphisms. We marked the Pan-Asian ethnic groups based on a maximum-likelihood tree of populations. Abbreviations are explained in Supplementary Table 1.
References
1. Cooper GM, Mefford HC. Detection of copy number variation using SNP genotyping. Methods Mol Biol 2011;767:243ŌĆō252. PMID: 21822880.


2. Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, et al. A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet 2007;80:91ŌĆō104. PMID: 17160897.



3. Claw KG, Tito RY, Stone AC, Verrelli BC. Haplotype structure and divergence at human and chimpanzee serotonin transporter and receptor genes: implications for behavioral disorder association analyses. Mol Biol Evol 2010;27:1518ŌĆō1529. PMID: 20118193.



4. Cordell HJ, Darlay R, Charoen P, Stewart A, Gullett AM, Lambert HJ, et al. Whole-genome linkage and association scan in primary, nonsyndromic vesicoureteric reflux. J Am Soc Nephrol 2010;21:113ŌĆō123. PMID: 19959718.



5. Moen T, Hayes B, Nilsen F, Delghandi M, Fjalestad KT, Fevolden SE, et al. Identification and characterisation of novel SNP markers in Atlantic cod: evidence for directional selection. BMC Genet 2008;9:18. PMID: 18302786.



6. Latter BD. Selection in finite populations with multiple alleles. 3. Genetic divergence with centripetal selection and mutation. Genetics 1972;70:475ŌĆō490. PMID: 5024717.


7. Hong H, Xu L, Liu J, Jones WD, Su Z, Ning B, et al. Technical reproducibility of genotyping SNP arrays used in genome-wide association studies. PLoS One 2012;7:e44483. PMID: 22970228.



8. McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet 2007;39(7 Suppl):S37ŌĆōS42. PMID: 17597780.


9. Kehrer-Sawatzki H, Cooper DN. Copy number variation and disease: preface. Cytogenet Genome Res 2008;123:5ŌĆō6. PMID: 19287133.


10. Schrider DR, Hahn MW. Gene copy-number polymorphism in nature. Proc Biol Sci 2010;277:3213ŌĆō3221. PMID: 20591863.



11. HUGO Pan-Asian SNP Consortium. Abdulla MA, Ahmed I, Assawamakin A, Bhak J, Brahmachari SK, et al. Mapping human genetic diversity in Asia. Science 2009;326:1541ŌĆō1545. PMID: 20007900.


12. Ngamphiw C, Assawamakin A, Xu S, Shaw PJ, Yang JO, Ghang H, et al. PanSNPdb: the Pan-Asian SNP genotyping database. PLoS One 2011;6:e21451. PMID: 21731755.



13. Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet 2006;78:629ŌĆō644. PMID: 16532393.



14. Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, et al. Large-scale copy number polymorphism in the human genome. Science 2004;305:525ŌĆō528. PMID: 15273396.


15. Park J, Hwang S, Lee YS, Kim SC, Lee D. SNP@Ethnos: a database of ethnically variant single-nucleotide polymorphisms. Nucleic Acids Res 2007;35:D711ŌĆōD715. PMID: 17135185.



16. Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A 2002;99:6567ŌĆō6572. PMID: 12011421.



18. Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 2011;39:D52ŌĆōD57. PMID: 21115458.



19. Kantola AK, Ryyn├żnen MJ, Lhota F, Keski-Oja J, Koli K. Independent regulation of short and long forms of latent TGF-beta binding protein (LTBP)-4 in cultured fibroblasts and human tissues. J Cell Physiol 2010;223:727ŌĆō736. PMID: 20175115.

20. Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 2009;459:569ŌĆō573. PMID: 19404257.



Fig.┬Ā1
Distribution of ethnic variant single nucleotide polymorphisms (A) and ethnic variant copy number polymorphisms (B) across ethnic groups. PG8 consists of Indo-European and Dravidian Southwest Asians. IN-NI, Mongoloid features; IN-TB, Mongoloid features; TH-MA, Mlabri; MY-MN, Malay; ID-ML, Malay; ID-LE, Lembata; PI-UI, Filipino; ID-DY, Dayak; PI-UN, Filipino; ID-SB, Kambera; IN-NL, Caucasoids; CN-UG, Uyghur; IN-EL, Caucasoids; IN-IL, Caucasoids; IN-SP, Caucasoids; JP-RK, Ryukyuan.
Table┬Ā1.
ESNP- and ECNP-related gene set summary
- TOOLS
-
METRICS

-
- 1 Crossref
- 6,880 View
- 54 Download





