Forensic Body Fluid Identification by Analysis of Multiple RNA Markers Using NanoString Technology
Article information
Abstract
RNA analysis has become a reliable method of body fluid identification for forensic use. Previously, we developed a combination of four multiplex quantitative PCR (qRT-PCR) probes to discriminate four different body fluids (blood, semen, saliva, and vaginal secretion). While those makers successfully identified most body fluid samples, there were some cases of false positive and negative identification. To improve the accuracy of the identification further, we tried to use multiple markers per body fluid and adopted the NanoString nCounter system instead of a multiplex qRT-PCR system. After measuring tens of RNA markers, we evaluated the accuracy of each marker for body fluid identification. For body fluids, such as blood and semen, each body fluid-specific marker was accurate enough for perfect identification. However, for saliva and vaginal secretion, no single marker was perfect. Thus, we designed a logistic regression model with multiple markers for saliva and vaginal secretion and achieved almost perfect identification. In conclusion, the NanoString nCounter is an efficient platform for measuring multiple RNA markers per body fluid and will be useful for forensic RNA analysis.
Introduction
RNA analysis has emerged as a reliable method of body fluid identification for forensic use [1-7]. Conventionally, immunological, enzymatic, and chemical detection of specific protein markers is used [8-10]. For example, prostate-specific antigen has been used for semen and hemoglobin for blood identification [9, 10]. These assays provide important information for crime scenes, but they can provide wrong information because of cross-reactions. Recently, several body fluid-specific mRNA markers have been discovered [1-7]: β-spectrin (SPTB), porphobilinogen deaminase (PBGD), and hemoglobin alpha locus 1 (HBA1) for blood; matrix metalloproteinase 7 and 11 (MMP7 and MMP11) for menstrual blood; statherin (STATH) and histatin 3 (HTN3) for saliva; kallikrein 3 (KLK3) and protamine 1 and 2 (PRM1 and PRM2) for semen; and human beta-defensin 1 (HBD-1) and mucin 4 (MUC4) for vaginal secretion [11]. After DNA/RNA co-extraction methods were applied to the forensic field, RNA analysis has become routine forensic analysis [12, 13].
RNA expression level is generally measured using reverse-transcription and standard end-point PCR (RT-PCR) or quantitative PCR (qRT-PCR) in a forensic lab. Recently, using microarray platforms, body fluid-specific mRNA markers were identified at a genome-wide level [14-16], and multiplex qRT-PCR probes of these markers were developed for one-step identification of body fluid type [7, 16]. Currently, most of the multiplex qRT-PCR assays use a single marker for each body fluid. However, using a single marker for each body fluid may lead to false positive/negative identification, as some markers are expressed in more than two types of body fluid [4-6]. Recently, Roeder and Haas [17] suggested a novel approach of using a minimum of five mRNA markers for each body fluid and a scoring method based on multiple markers. In this regard, the multiplex qRT-PCR system is not useful, because the number of fluorescent dyes in one reaction is limited.
The NanoString nCounter (NanoString Technologies, Seattle, WA, USA) is a recent platform that can quantify the expression of hundreds of mRNAs in a single reaction using color-coded molecular barcodes [18, 19]. NanoString nCounter is also known as a sufficiently robust method to measure expression in degraded RNA samples, such as formalin-fixed paraffin-embedded tissues and crude tissue lysates [18, 20]. Therefore, applying NanoString nCounter to forensic identification of body fluids enables the digital quantification of multiplexed markers for each body fluid.
In this study, we adopted the NanoString nCounter and designed a panel of NanoString probes for multiple markers for each body fluid. We tested the multiple markers in a large number of body fluid samples. As a result, we found that the new multiplexed method could improve the specificity and sensitivity of identification. We suggest that using multiple mRNA markers for each body fluid could improve the accuracy of body fluid identification.
Methods
Sample collection and RNA preparation
Twelve samples for each body fluid (blood, saliva, semen, and vaginal secretion) were collected from healthy Korean volunteers with informed consent from the participants. The study protocol was approved by the Institutional Review Boards of Chungnam National University Hospital. To prepare total RNA, we employed the Qiagen RNeasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Extracted total RNA was analyzed using Experion RNA StdSens (Bio-Rad, Hercules, CA, USA) to check its quality and quantity.
NanoString experiment
To validate the mRNA candidates as body fluid specific markers, NanoString technology was employed. Color-coded barcodes that represented a single target mRNA were synthesized, targeting 18 body fluid-specific mRNA markers, and 2 endogenous controls for mRNA. Briefly, 100 ng of total RNA was hybridized to the barcode, and then probe-mRNA complexes were immobilized on a streptavidin-coated cartridge according to the manufacturer's protocol. Subsequently, the cartridges were placed in the digital analyzer, and barcodes were counted. All the counts were normalized by the count of GAPDH barcodes.
Statistical analysis
We applied student's t-test to evaluate the significance of gene expression differences between the tissue of interest and the other three tissues among the four body fluids. We applied receive operating characteristic (ROC) analysis to estimate the sensitivity and specificity of each marker, using the ROCR package [21] of R software (version 2.6.1). Again, the four body fluids were divided into two groups: a tissue of interest and the remaining three tissues among the four body fluids. Results with a p-value of < 0.05 were considered significant.
Results
Previously, we found a dozen mRNA markers and developed multiplex qRT-PCR probes for body fluid identification [16]. But, most of the markers, except pro-platelet basic protein (PPBP), were not perfect in terms of area under the curve (AUC) value (AUC, 1). To improve the accuracy of identification, we considered a multiplexed assay using multiple probes for each body fluid. While the multiplex qRT-PCR system is limited by the number of available dyes in a reaction, the NanoString nCounter can measure the expression of hundreds of RNAs at once [18]. So, we adopted the NanoString nCounter as the multiplexed identification method using multiple RNA markers for each body fluid.
After designing NanoString probes for 18 mRNA markers (four to five for each body fluid and two for controls), we performed NanoString nCounter assays with a total of 12 RNA samples for each body fluid. As a result, we got a dozen good probes (Fig. 1) according to the following criteria: 1) p < 0.005 and 2) body fluid-specific expression pattern. Results of the other probes are shown in Supplementary Fig. 1. To test the value of each marker as a body fluid-specific marker, we performed ROC analysis (Fig. 2). For blood- or semen-specific markers, the identification accuracy was perfect (AUC, 1). But, for saliva and vaginal secretion, no single marker showed perfect accuracy (Fig. 2).

Expression patterns of selected mRNA markers in our NanoString nCounter assay dataset. Each count was normalized to GAPDH mRNA. (A) Blood. (B) Saliva. (C) Semen. (D) Vaginal.

Receive operating characteristic analysis of selected mRNA markers in our NanoString nCounter assay dataset. AUC, area under the curve. (A) Blood. (B) Saliva. (C) Semen. (D) Vaginal.
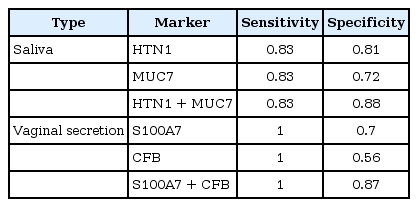
To improve the accuracy of saliva and vaginal secretion identification, we tried a combination of markers using logistic regression analysis [22]. We evaluated the sensitivity and specificity of each marker and then evaluated the performance of the combined markers (Fig. 3). For saliva and vaginal identification, using two markers significantly improved the identification accuracy (Table 1). Therefore, we could identify each body fluid with multiple makers for each body fluid using the NanoString nCounter quite accurately.
Discussion
Previously, we developed multiplex qRT-PCR probes that used one specific mRNA marker for each body fluid [16]. In that work, blood was perfectly identified using a single marker, PPBP. However, for other body fluids, such as vaginal secretion and saliva, it was difficult to find out markers that perfectly discriminated them from other body fluids. For example, as saliva and vaginal secretion showed similar expression patterns, many selected body fluid-specific markers were expressed in both body fluids. This problem led us to consider a combination of multiple markers for perfect identification.
A limit in the number of markers that can be assessed in a reaction was the biggest problem when we considered the use of multiple markers for each body fluid. As the number of different fluorescent dyes available in a qRT-PCR reaction is limited (i.e., five), we considered a new method that can measure at least 10 RNAs at once. Fortunately, the recently developed NanoString nCounter allowed us to measure hundreds of probes in a single reaction; so, we adopted the NanoString platform in our body fluid identification project.
Measuring the expression of multiple makers per body fluid has a big advantage. As we have shown, by using two markers for each body fluid, we could identify saliva and vaginal secretions that were not identified perfectly by a single marker. As we could identify blood and semen perfectly using only one marker per body fluid, we conclude that a total of 10 markers is enough to identify four different body fluids in one reaction. In this regard, the NanoString nCounter is a promising platform that allows us to identify each body fluid in one reaction with high accuracy.
Acknowledgments
This work was supported by the Forensic Science Research Project 2013 of the Supreme Prosecutors' Office and a KRIBB Research Initiative Grant.
References
Supplementary material
Supplementary data including one figure can be found with this article online at http://www.genominfo.org/src/sm/gni-11-277-s001.pdf.
Supplementary Information
Forensic Body Fluid Identification by Analysis of Multiple RNA Markers Using NanoString Technology