Genes Involved in the Biosynthesis and Transport of Acinetobactin in Acinetobacter baumannii
Article information
Abstract
Pathogenic bacteria survive in iron-limited host environments by using several iron acquisition mechanisms. Acinetobacter baumannii, causing serious infections in compromised patients, produces an iron-chelating molecule, called acinetobactin, which is composed of equimolar quantities of 2,3-dihydroxybenzoic acid (DHBA), L-threonine, and N-hydroxyhistamine, to compete with host cells for iron. Genes that are involved in the production and transport of acinetobactin are clustered within the genome of A. baumannii. A recent study showed that entA, located outside of the acinetobactin gene cluster, plays important roles in the biosynthesis of the acinetobactin precursor DHBA and in bacterial pathogenesis. Therefore, understanding the genes that are associated with the biosynthesis and transport of acinetobactin in the bacterial genome is required. This review is intended to provide a general overview of the genes in the genome of A. baumannii that are required for acinetobactin biosynthesis and transport.
Introduction
Iron is one of the essential nutrients for host and pathogenic bacteria, and both require the metal as a cofactor or as a prosthetic group for biologically important proteins that are involved in many basic cellular functions [1]. Most iron in the mammalian host is sequestered and bound to high-affinity iron-binding proteins, such as transferrin, lactoferrin, and ferritin, or complexed to the heme of hemoproteins [2]. Hence, iron is rarely found as free iron in the mammalian host and is one of the major limiting factors of pathogenic bacteria. Therefore, pathogenic bacteria have evolved sophisticated mechanisms for the acquisition of iron from host tissues because of the limited availability of iron to the bacteria and the inevitable competition with the host [3, 4, 5, 6, 7, 8].
Siderophores are high-affinity iron-chelating compounds secreted by pathogenic bacteria [3]. Siderophores are classified into three major groups-catecholates, hydroxamates, and carboxylates-by the ligands that are used to chelate the ferric iron [7]. The diversity of siderophores may have resulted from evolutionary pressure, causing the bacteria to produce structurally different siderophores that can not be transported by specific transport systems of other bacteria. For example, Escherichia coli produces a prototype catechol siderophore, which is a cyclic trimer of 2,3-dihydroxybenzoyl-L-serine [5]. In comparison, vibriobactin, produced by Vibrio cholera, is a non-cyclic derivative of 2,3-dihydroxybenzoic acid (DHBA) [9]. Genes that are involved in the biosynthesis and uptake of siderophores are often clustered within the bacterial genome, and their products are closely linked to the virulence of pathogenic bacteria.
Like the pathogenesis of many other bacteria, the pathogenesis of Acinetobacter baumannii, a major opportunistic pathogen that causes serious diseases in compromised patients, including pneumonia, bacteremia, urinary tract infection, sepsis, and meningitis [10, 11], depends primarily on the organism's ability to uptake and utilize iron [12, 13, 14]. Numerous studies have investigated the response of various A. baumannii isolates in iron-limited conditions and identified several gene clusters that are required for the synthesis and transport of siderophores [15]. Especially, the acinetobactin-mediated iron acquisition system has been the most extensively characterized system in A. baumannii [16, 17]. Acinetobactin, a catechol-hydroxamate siderophore, which is a non-cyclic derivative of DHBA that is linked to threonine and N-hydroxyhistamine, is produced and utilized by three putative systems encoded within the acinetobactin gene cluster of A. baumannii. Moreover, A. baumannii strains have significant genetic diversity in the numbers and types of these iron uptake and utilization systems [15]. Therefore, these results indicate that genetic diversity in acinetobactin gene clusters could be a major factor in determining the virulence among A. baumannii strains. In this review, we describe a gene cluster in the genome of A. baumannii that is involved in the biosynthesis and transport of acinetobactin. Furthermore, we present the genetic diversity of the gene cluster that is responsible for acinetobactin biosynthesis and transport in different A. baumannii strains.
Acinetobactin Gene Cluster in A. baumannii
A. baumannii express high-affinity iron acquisition systems, such as siderophores, to survive under iron-limited host environments. The iron acquisition system that is mediated by the siderophore acinetobactin has been extensively characterized in A. baumannii. Especially, the gene cluster involved in the biosynthesis and transport of acinetobactin has been found in various clinical isolates of A. baumannii, except A. baumannii SDF [15]. Acinetobactin is synthesized from a DHBA, threonine, and hydroxyhistamine via proteins encoded by genes within the gene cluster. The mixed-type siderophore, which consists of catecholate and hydroxamate groups, exhibits high affinity for iron [16, 17]. Particularly, it has been found that 18 coding regions in the chromosome of A. baumannii ATCC 19606 are required for acinetobactin biosynthesis and the uptake of ferric-acinetobactin complexes [15, 18]. The acinetobactin gene cluster of A. baumannii ATCC 19606 is illustrated in Fig. 1. The acinetobactin-mediated iron acquisition system is composed of three putative systems encoded within the acinetobactin cluster. First, the biosynthesis of acinetobactin is accomplished by the proteins encoded by basA, basB, basC, basD, basF, basG, basH, basI, and basJ [14, 15]. Second, the acinetobactin that is produced by A. baumannii is secreted via a siderophore efflux system of the ABC superfamily, consisting of the proteins encoded by barA and barB [18]. Finally, ferric-acinetobactin complexes move into bacterial cells via a receptor for ferric-acinetobactin complexes, consisting of the proteins encoded by bauA, bauB, bauC, bauD, bauE, and bauF [14]. Especially, bauB, bauC, bauD, and bauE play important roles in the translocation of ferric-acinetobactin complexes into bacterial cells [15]. In addition, putative binding sites for ferric uptake regulator, which controls bacterial iron homeostasis, exist in the upstream regions of basJ, entE/basD, basA/bauF, and bauD/basD. All of these genes that are involved in the biosynthesis and transport of acinetobactin are significantly up-regulated under iron-limited conditions [15]. Particularly, a functional analysis of basD, bauA, and bauD mutants under iron-limited conditions showed that iron acquisition that is mediated by the biosynthesis and transport of acinetobactin is essential for the survival of A. baumannii under iron-limited conditions [13].

Physical map of siderophore gene clusters in Acinetobacter baumannii. Siderophore gene clusters in different A. baumannii strains. The gene clusters are based on the database of A. baumannii ATCC 19606, AB0057, ACICU, and AB307-294, which were retrieved separately from GenBank (accession Nos. ACQB00000000, CP001182-3, CP000863, and CP001172). Each color indicates the biological functions in the biosynthesis and transport of acinetobactin. Red arrows represent the genes that were added in the acinetobactin gene cluster.
Moreover, a recent study by Gaddy et al. [13] indicated that BasD and BauA play important roles in the persistence of infection by A. baumannii within epithelial cells and their apoptotic death. The results obtained from a mouse sepsis model also showed that the acinetobactin-mediated iron acquisition system is a major factor in the pathogenesis of A. baumannii, such as the establishment of bacterial infection and cell damage. Especially, impairment of BasD production resulted in an intermediate virulence phenotype in ex vivo and in vivo infection models in comparison to those of the parental strain and bauA mutant. These results represent that both acinetobactin synthesis and uptake of iron-acinetobactin complexes are absolutely required for the pathogenesis of A. baumannii.
Genetic Diversity of Acinetobactin Gene Cluster in Different A. baumannii Strains
Penwell et al. [18] described that the gene cluster that is involved in acinetobactin production and its utilization exists in the chromosomes of different A. baumannii strains. In addition, the gene cluster is found in A. baumannii AYE, which is a natural entA mutant incapable of producing acinetobactin [18, 19]. However, the cognate clusters in the A. baumannii AB0057 and ACICU strains contain the additional putative genes AB57_2807 and AB57_2818 [20], and ACICU_02575 and ACICU_02586 [21], respectively (Fig. 1). In the case of A. baumannii AB307-294, three additional predicted genes, ABBFA_001054, ABBFA_001053, and ABBFA_001065, are also added in the bas-bau-bar gene cluster (Fig. 1) [20]. Although the functions of these additional genes in siderophore production and its utilization are not characterized, these results indicate the possibility that genetic diversity in acinetobactin gene clusters could be closely linked to the virulence of A. baumannii.
entA Ortholog in A. baumannii
Enterobactin that is produced by E. coli is a high-affinity siderophore for iron [5]. In the enterobactin biosynthesis, chorismic acid is converted to DHBA by the products of entA, entB, and entC. An amide linkage of DHBA to L-serine (Ser) is then catalyzed by the proteins encoded by entD, entE, entF, and entB. The molecules of the DHBA-Ser that is formed undergo intermolecular cyclization, yielding enterobactin. A. baumannii also employs basF and basJ within the acinetobactin gene cluster, which are homologs of entC and entB, respectively, to produce mature acinetobactin [18]. However, entA has not been found in the acinetobactin gene cluster. Recently, it was found that a single functional copy of an entA ortholog is located in anupstream region of the modE-modA-modB-modC gene cluster that is required for the molybdenum transport system in A. baumannii ATCC 19606 (Fig. 2) [18]. In particular, a functional analysis based on gene cloning and biochemical tests showed that entA is absolutely required for the production of DHBA, which is used by A. baumannii as a precursor for acinetobactin biosynthesis. The study also demonstrated that acinetobactin intermediates and DHBA play important roles in the virulence of A. baumannii in ex vivo and in vivo infection models.
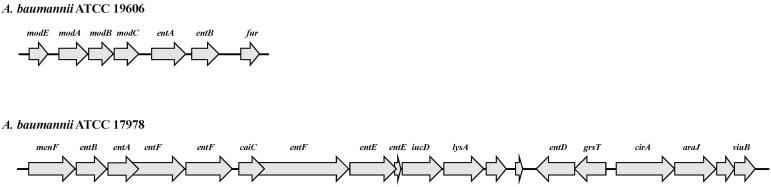
Genetic map of the gene cluster containing the entA ortholog. The arrows represent the coding regions of the genes identified in the genomes of Acinetobacter baumannii ATCC 19606 and ATCC 17978. The names of the genes are shown above each coding region.
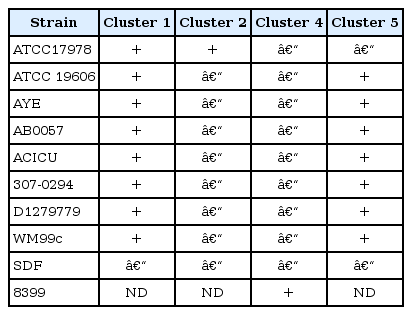
Even though entA is found outside of the acinetobactin cluster in A. baumannii strains, the genetic contents surrounding the entA gene are variable among different clinical isolates [18]. For example, the entA gene of A. baumannii ATCC 19606 is next to the coding regions of the mod gene cluster (Fig. 2). However, the entA gene of A. baumannii ATCC 17978 is situated within a large gene cluster that encodes an uncharacterized siderophore-mediated system (Fig. 2). Especially, this cluster, in which mobile DNA elements exist, is flanked by inverted repeats, a feature that can explain its transfer by lateral processes. Therefore, these results represent that genetic components that are required for iron acquisition via the acinetobactin-mediated system give rise to horizontal gene transfer with complex chromosomal rearrangement processes. Consequently, the newly added siderophore gene cluster with the entA gene via horizontal gene transfer may cause these two strains, ATCC 19606 and ATCC 17978, to acquire iron via different siderophore-mediated systems from different sources. The possibility was proven by a genomic analysis of fully sequenced and annotated A. baumannii isolates [15, 19]. Four siderophore gene clusters are newly predicted to be involved in siderophore production and the uptake of iron-siderophore complexes (Table 1). Cluster 1 is indicated in the genomes of all strains analyzed, with the exception of A. baumannii 8399. Cluster 2 and cluster 4 are found only in A. baumannii ATCC 17978 and isolate 8399, respectively. Cluster 5 is present in all genomes analyzed except for that of A. baumannii ATCC 17978. The possibility is also supported by the result that the iron uptake phenotype of the A. baumannii ATCC 17978 basD mutant was not different from that of the parental 17978 strain when compared under iron-limited conditions.
Conclusion
A. baumannii employs specialized systems that are required for iron acquisition in iron-limited host environments [16]. In this review, we described the genes of A. baumannii that are involved in acinetobactin biosynthesis and transport. A number of studies have identified the genes that are essential for iron acquisition [13, 15, 16, 17]. Especially, the data presented in this review indicate that A. baumannii strains have genetic diversity in the acinetobacter gene cluster. Even though the roles of the additional genes in acinetobactin cluster in different. A. baumannii strains are not characterized, we suggest that the genes could be associated with A. baumannii virulence. Therefore, functional analysis of the genes in the bacteria is required. Furthermore, we showed both a single entA functional ortholog and variability in the bacterial genome. Therefore, we suggest that A. baumannii has continuously evolved the iron acquisition mechanism, which is associated with acinetobactin from different sources by horizontal gene transfer.
Acknowledgments
This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI14C0257).