 |
 |
- Search
| Genomics Inform > Volume 11(1); 2013 > Article |
Abstract
As the International Human Epigenome Consortium (IHEC) launched officially at the 2010 Washington meeting, a giant step toward the conquest of unexplored regions of the human genome has begun. IHEC aims at the production of 1,000 reference epigenomes to the international scientific community for next 7-10 years. Seven member institutions, including South Korea, Korea National Institute of Health (KNIH), will produce 25-200 reference epigenomes individually, and the produced data will be publically available by using a data center. Epigenome data will cover from whole genome bisulfite sequencing, histone modification, and chromatin access information to miRNA-seq. The final goal of IHEC is the production of reference maps of human epigenomes for key cellular status relevant to health and disease.
Epigenome is the word describing the thing that is not included in genetics. "Epi" means upon or outside; so, a direct translation is "the thing that is not related with genetics or above genetics." Practically, it means gene regulation phenomena, including by means of DNA methylation, histone modification, and miRNA. To explore epigenomic regulation, International Human Epigenome Consortium (IHEC) was initiated to profile the epigenome of all human cell types. To understand the current IHEC, it is required to turn back into the initial steps for fostering this consortium. It has passed more than a decade after completion of the human genome project, and several efforts were conceived to accelerate finding disease-causing genetic variations, such as genome-wide association study (2003), Encyclopedia of DNA Element (ENCODE, 2003), The Cancer Genome Atlas (TCGA, 2006), International Cancer Genome Consortium (ICGC, 2008), and National Institutes of Health (NIH) Roadmap Epigenomics (2008) (Table 1). IHEC was born with ground making effort of these several international consortiums. Although the Human Genome Project, which defined the exact sequence of 3 billion bases in DNA, is one of the big milestones in genome research, cataloging DNA sequence information has a limited effect on defining whole varieties of genome variations that are expected to be crucial cause of disease. Also, the sequence itself does not provide how the genome is packaged in chromosomes and chromatin, which is essential for organismal development and differentiation. So, epigenetic regulation, in addition to transcription factor binding profile, is emerging as a powerful tool to identify disease correlation with genome information [1]. In 2004, a National Cancer Institute (NCI)-sponsored workshop, Epigenetic Mechanisms in Cancer, had initiating discussions of necessity of the Human Epigenome Project, which will be in parallel with the Human Genome Project. In 2005, there was a special workshop on the BLUEPRINT for a Human Epigenome Project, the American Association of Cancer Research (AACR) Human Epigenome Workshop, on defining human epigenomes at high resolution.
The consensus of workshop participants was that there were compelling reasons from both scientific and public health perspectives to initiate a human epigenome project that could take full advantage of advances in several existing US and European initiatives. In 2006, TCGA began as a 3-year pilot with an investment of $50 million each from the NCI and National Human Genome Research Institute (NHGRI) to analyze the genome and epigenome of 20 types of cancer tissue. In 2008, amending the TCGA, international efforts were assembled to analyze cancer genome/epigenome variation. As a result, the ICGC was launched in 2008 to coordinate large-scale cancer (epi)genome studies in tumors from 50 cancer types and/or subtypes, with participation of 15 countries. ICGC is focused on obtaining a comprehensive description of genomic, transcriptomic, and epigenomic changes in 50 different tumor types. Also, the international Alliance for Human Epigenetics and Disease (AHEAD) [2] was conceived among participating countries. As a result, the Roadmap Epigenomics Program was fostered as AHEAD's suggestion. Finally the NIH Roadmap Epigenomics Program was initiated with the funding of $200 million for 5 years from NIH [3]. Roadmap Epigenomics aimed to produce reference epigenome data from a variety of human cells. As NIH and European epigenome mapping activity was commenced as a locally scattered project, there were efforts to assemble reference epigenome mapping as an international collaboration. From 2009, several workshops were held in NIH or Europe, and finally at the 2010 Washington meeting, IHEC was officially born.
Through February 2013, 7 member organizations/countries joined IHEC, including NIH Roadmap Epigenomics, EU BLUEPRINT, Germany Deutsches Epigenom Programm (DEEP), Canadian Institutes for Health Research (CIHR), Japan Science and Technology Agency (JST), Korea KNIH, and Italy. Several other programs, such as ENCODE, England, France, and Singapore, expressed their intents to join IHEC. IHEC consists of an Executive Committee (EXEC, association of funding agency) and International Scientific Steering Committee (ISSC, representatives of the member research institution).
IHEC aims at cataloging 1,000 human reference epigenomes for the next 7 to 10 years. The 1,000 epigenome includes 250 types of human cells, and 10% of the epigenome can be mapped on a model organism. A long-term objective of IHEC is to determine the extent to which the epigenome has shaped human populations over generations and in response to the environment and to produce human reference epigenome maps for key cellular status relevant to health and disease. IHEC will focus on key cellular status, such as stemness, immortality, proliferation, differentiation, senescence, and stress, thereby generating new knowledge that will catalyze progress in health research and regenerative medicine. IHEC will coordinate rapid distribution of the data to the entire research community with minimal restrictions to accelerate translation of this new knowledge into health and diseases. IHEC also will coordinate the development of common bioinformatics standards, data models, and analytical tools to organize, integrate, and display whole epigenomic data generated from this important international effort. IHEC will set up an efficient structure that will coordinate this international effort so that the interest and priorities of individual participants, self-organizing consortia, funding agencies, and nations are addressed. IHEC will encourage the minimal amount of redundancy between the different epigenetics efforts around the world. IHEC will also interact and coordinate its efforts with other international projects, such as ICGC and ENCODE. The secondary goal of IHEC is to catalyze the development of new and robust technologies that will facilitate the characterization and functional analysis of the epigenome in health and diseases, thereby driving down the costs of epigenome mapping substantially, and to support the dissemination of knowledge and standards related to new technologies, software, and methods to facilitate data integration and sharing between epigenetics researchers around the globe. By the production of 1,000 epigenomes, IHEC expects that we can understand the key cellular aspects of 250 types of human cells more deeply. Also, having ~250 types of reference epigenomes, we can have the power to compare disease epigenomes to normal references, which will help explain the missing heritability of common complex diseases. Now, the production of >600 epigenomes has been planned, and ~60 epigenomes have been finished [3, 4].
The NIH Roadmap Epigenomics Program began in 2008 and has a budget of $200 million. Under the cover of this program, the NIH Common Fund and NIH Institutes and Centers have supported a total of 68 grants in the areas of epigenetic technology development, identification of novel epigenetic marks, reference epigenome mapping, and disease epigenomics investigations. The Reference Epigenomic Mapping Consortium, funded through the Common Fund's Roadmap Epigenomics Program, has been generating genome-wide epigenomic maps for a variety of cell and tissue types. The majority of reference epigenomes that have been generated contains information on epigenetic modifications, including a core set of histone marks, DNA methylation, chromatin accessibility, and gene expression information. A subset of reference epigenomes also contains an expanded set of 20 additional histone modifications [5]. Data for 52 complete and partial epigenome datasets for varieties of normal human cells and tissues are available. Epigenomes were mapped on embryonic stem (ES) cells, ES cell derivatives, induced pluripotent stem cells, multiple fetal tissues, several varieties of blood and immune cells, breast cell types, placenta, and solid tissues (e.g., adipose, gastrointestinal tract, skin, and brain). Roadmap Epigenomics plans to complete ~100 additional epigenomes by the end of the program [3, 4].
The BLUEPRINT consortium, consisting of 41 leading European universities, research institutes, and industry entrepreneurs, was selected for funding in early 2011. The project officially started on October 1, 2011 and will run for 4-5 years. BLUEPRINT has an overall budget of 39.4 million €, for which it receives 30 million € in funding from the European Commission. BLUEPRINT aims to generate at least 100 reference epigenomes on blood lineage (hematopoietic) cells from healthy individuals and on their malignant leukemic counterparts.
The project proposal from KNIH, the Center for Genome Sciences, Epigenome Mapping Center (EMC), 5 chronic disease-related hospital, were finally selected for funding during the period of 2012-2013. The project officially started on January 2012 and will run for >5 years. KNIH has an overall budget of $10.9 million for 5 years. Participating clinical institutions were chosen from Samsung Medical Center (heart tissue and cardiomyopathy), Hanyang University Hospital (blood, synoviocyte cell, and rheumatoid arthritis), Asan Medical Center (pancreatic tissue, adipose tissue, T2D, and obesity), and Seoul National University Hospital (kidney tissue and chronic kidney disease). KNIH aims to generate at least 50 reference epigenomes and study them to advance and exploit knowledge of the underlying biological processes and mechanisms in health and disease. KNIH will focus on 50 distinct types of homogenous cells of tissues related to 5 chronic diseases (heart failure, autoimmune diseases, diabetes, obesity, and chronic kidney disease) from normal and disease tissue, mainly obtained by tissue removal or transplantation.
The Korea reference epigenome will entail the analysis of 12 'marks'; an IHEC CORE set: DNAme, H3K4me3, H3K4me1, H3K9me3, H3K27me3, H3K27ac, H3K36me3, RNA-seq, and RNA-chromatin immunoprecipitation (RNA-ChIP), DNaseI hypersensitivity, formaldehyde-assisted isolation of regulatory elements (FAIRE)-seq, chromatin interaction analysis using paired end tags (CHIA-PET), and miRNA-seq (Tables 2 and 3) [4].
JST's leading program is the Core Research for Evolutional Science and Technology (CREST)'s disease epigenome project. In 2011, JST started a research area of "Development of fundamental technologies for diagnosis and therapy based upon epigenome analysis (disease epigenome)." This research area focuses on the IHEC projects and various epigenome analyses related to diseases and stem cells. In 2011-2012, 10 projects were awarded, including 3 IHEC projects. JST' proposal focused the epithelial cells of the digestive system (stomach, colon, and liver), the vascular endothelial cells, and the epigenome in germ cells and reproductive tissue (placental cell, cytotrophoblast, syncytiotrophoblast, endometrial cells, basal layer cells, proliferative functional layer cells, and secretary functional layer cells) (Tables 2 and 3) [4].
The Italian epigenome program involves two initiatives: the Milan Genomic Initiative (MGI) and the Italian Flagship Project EPIGEN. The MGI was started in 2011 as a joint operation among Istituto Europeo di Oncologia (IEO), Istituto Italiano di Technologia (IIT), and Istituto FIRC di Oncologia Molecolare (IFOM) and is located at the IFOM-IEO campus in Milan, a multi-institutional biomedical research area hosting over 500 scientists. The MGI includes 16 independent research-groups in the field of epigenetics. The three institutions are committed to 5-year support of the "Center of Genomic Science" for a total of ~10 million €. EPIGEN was launched by the Italian Ministry of Research and started in January 2012. It includes 25 independent groups. EPIGEN is supported by funding of 30 million € for 4 years. MGI has been a member of IHEC since 2011, and EPIGEN is currently following the required procedures with the Italian Ministry of Research for formal application to IHEC. In parallel, both initiatives are working on a coordination plan with the goal of joint participation to IHEC. MGI expects to provide 50 reference epigenomes, completed to IHEC standards, over the next 5 years, including cells from hematopoietic, mammary, adipose, liver, and neural tissues. Epigen focuses on research projects, such as the role of chromatin structure, DNA methylation, and nuclear architecture in epigenetic regulation and epigenetic control through the noncoding component of the genome: structure and dynamics, epigenetic control of cellular identity and human pathologies, epidrugs for therapeutic use, and plant epigenomics [4].
DEEP's funding is approx. 16 million € by the German Federal Ministry of Education and Research (BMBF), from 2012 to 2017. DEEP creates an interdisciplinary research platform of 17 subprojects, linking 21 epigenomic mapping and functional analysis groups across Germany. As the German contribution to IHEC, DEEP plans to produce and functionally interpret 70 reference epigenomes of selected human (and some murine) cells and tissues. DEEP will focus on cell types connected to metabolic diseases, such as steatosis (hepatocytes and Kupffer cells) and obesity (adipocytes [small/large, visceral/subcutaneous], monocytes, and macrophages), as well as inflammatory diseases, such as Crohn disease (mucosa, macrophages, and T-memory/effector cells) and rheumatoid arthritis (fibroblasts, macrophages, and T-memory/effector cells) (Tables 2 and 3) [6].
In 2011, the CIHR established a national collaborative research funding strategy to coordinate epigenetics research in Canada, CEEHRC. The budget of the CEEHRC is $50 million over 7 years. CEEHRC is supporting two EMCs and two Epigenomic Data Coordination Centers (EDCCs), for a total investment of $15 million over 5 years. The CEEHRC-funded EMCs are the Center for Epigenomic Mapping Technologies (CEMT), at the University of British Columbia/BC Cancer Agency, and the Multidimensional Epigenomics Mapping Center at McGill (MEMCM), at McGill University. The funded EDCCs are at Simon Fraser University/BC Cancer Agency and at McGill University. The CEMT group is focusing on a number of cancer and stem cell types, including studies on peripheral and lymphoid B-cells, T-lymphocytes, primitive hematopoietic cells, and patient-derived induced pluoripotent stem cell, as well as thyroid cells and mammary, endometrial, fallopian, and colonic epithelium. The MEMCM group has three initial focus areas: autoimmune and inflammatory disease (using various blood cell types), cardio-metabolic disease (including cardiac and skeletal muscle, liver, adipose tissue, and pancreatic islets), and neuropsychiatric disease (including targeted brain areas in post mortem suicide human tissue, and animal models of hypothalamus/pituitary/adrenal axis dysfunction). In addition, MEMCM specifically addresses population variations in epigenomes within these focus areas. The EMCs together expect to provide 200 reference epigenomes, completed to IHEC standards, over the next 5 years (Tables 2 and 3) [6].
The normal phenotype of a sample should be macroscopically and microscopically confirmed by at least two pathologists. To monitor the cell quality, RNA quality should be assessed by RNA integrity number (RIN). The value is expected to exceed 8 (cell lines) or 4 (cells isolated from primary tissues) and should be described as the sample information. The purity of cells whose epigenome is analyzed must be assessed by immunocytochemistry, cell sorting, or other methods. The cell purity is expected to exceed 90%. However, if this is practically difficult, the purity of a sample should be described as the sample information. For optimal ChIP results, it is recommended to use more than 1 × 106 cells per chromatin immunoprecipitation.
The IHEC epigenome set consists of methylome (Bisulfite-seq [required], methylated DNA immunoprecipitation-seq [MeDIP-seq], methylation-sensitive restriction enzyme-seq [MRE-seq], reduced representation bisulphite-seq [RRBS], methylation capturing-seq [methylCap-seq] [any of 4 required on Roadmap]), RNA (RNA-seq, RNA-ChIP [required to Roadmap], smRNA-seq [optional]), ChIP-seq [input, H3K27me3, H3K36me3, H3K4me1, H3K4me3, H3K27ac, H3K9me3], and chromatin accessibility (DNaseI hypersensitivity, digital genomic footprinting, FAIRE-seq [any 3 optional]) (Table 3) [6].
Metadata standards are the data-related information required for data integration produced from individual IHEC institutions. It consists of 3 components: data standards, assay standards, and programmatic interoperability and programming interfaces, such as Representational State Transfer Application Programing Interfaces (REST APIs), allowing programs (apps) to pull data and metadata together from various sources across the web. Metadata standards allow computer programs and end users to access essential information about performed experiments, such as the information about assays, samples assayed, and algorithms used to produce interpretable data. In the preparatory stage, the project was to complete a case study in data and software integration using the Genboree system [6]. Several standards have yet to be defined, particularly controlled vocabularies and ontologies for important data elements, such as sample types, disease conditions, and phenotypes [6].
The IHEC data ecosystem project is to provide the infrastructure of data repositories and resources that enable data sharing by working together for the collection, processing, storage, and distribution of data from IHEC. Data are normally submitted to the central data repository site by individual data producers. Processed IHEC data, such as peak calls and epigenome maps, may not be deposited in a central data repository and may be only made available directly by the data distribution component (local) of the data ecosystem. There is value in distributing both the raw and processed data, and both will be provided. A virtual data integration center will be conceived in developing an IHEC data sharing system, and a joint project with a metadata standard project will be done in the near future [6, 7].
Although the international consortium is now in good shape, it still has several limitations. As the IHEC aims at producing 1,000 epigenomes from ~250 human cell types, there is a limitation on tissue or cell availability. Epigenome production from extremely hard-to-get cells or tissues seems impossible to accomplish and looks even unnecessary. To overcome this limitation, there are some efforts to perform ChIP-seq with fewer than ~1,000 cells and to perform single-cell-level epigenome analysis [14]. Also, there is an individual variation issue: as epigenomes differ by individual, age, cell type, and developmental stage, it will be very hard to determine the real reference data among numerous epigenomic states. It is required to determine the exact reference epigenome more precisely. Moreover, overlapping target tissues among member institutions is a limitation. Cell types, such as blood and adipocyte, that easy to get seem to be popular targets in every IHEC member and should be coordinated in a central way to foster synergy from each mapping activity. Another issue is a limitation of international collaboration. As all funds are controlled by an individual institution's decision, drawing a concrete conclusion on a specific point is expected to be a time-consuming process. There is newly emerging technology on targeting several novel epigenome sets, such as carboxyl methylation [15], m6A RNA methylation [16], hydroxyl methylation [8], and 3D chromosomal interaction [17]. Also, collaboration with the ENCODE project on transcription factor binding site profiling should be actively considered in near future (Fig. 1). Furthermore, in the next phase of IHEC, disease epigenome should be considered. Actually, all 7 member countries are focusing on disease relatedness actively, such as the metabolic epigenome, hematopoietic epigenome, and digestive organ disorders, but more active engagement and control should be anticipated in each project's detailed goals. Extension to epigenome wide association study should also be considered to increase the clinical importance of IHEC's reference mapping efforts [9].
Acknowledgments
This work was supported by grants from the Korea Centers for Disease Control and Prevention (4845-301, 4846-300) and an intramural grant from the Korea National Institute of Health (2012-N73006-00).
References
1. Jones PA, Martienssen R. A blueprint for a Human Epigenome Project: the AACR Human Epigenome Workshop. Cancer Res 2005;65:11241–11246. PMID: 16357125.


2. American Association for Cancer Research Human Epigenome Task Force. European Union, Network of Excellence, Scientific Advisory Board. Moving AHEAD with an international human epigenome project. Nature 2008;454:711–715. PMID: 18685699.




3. NIH roadmap epigenomics. Accessed 2013 Jan 30. Bethesda: National Center for Biotechnology Information, Available from: http://www.roadmapepigenomics.org.
4. International Human Epigenome Consortium (IHEC). International Human Epigenome Consortium Accessed 2013 Jan 30. Available from: http://www.IHEC-epigenomes.net.
5. Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 2010;28:1045–1048. PMID: 20944595.



6. Blueprint Epigenome. Accessed 2013 Jan 30. Nijmegen: Blueprint Epigenome, Available from: http://www.blueprint-epigenome.eu/.
7. Challis D, Yu J, Evani US, Jackson AR, Paithankar S, Coarfa C, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics 2012;13:8. PMID: 22239737.



8. Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 2012;150:1135–1146. PMID: 22980977.



9. Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet 2011;12:529–541. PMID: 21747404.



10. Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009;462:315–322. PMID: 19829295.



11. Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 2011;471:68–73. PMID: 21289626.



12. Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012;337:1190–1195. PMID: 22955828.



13. Sandhu KS, Li G, Poh HM, Quek YL, Sia YY, Peh SQ, et al. Large-scale functional organization of long-range chromatin interaction networks. Cell Rep 2012;2:1207–1219. PMID: 23103170.



14. Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 2010;7:461–465. PMID: 20453866.



15. Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol 2010;11:607–620. PMID: 20683471.



16. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 2012;149:1635–1646. PMID: 22608085.



17. Zhang J, Poh HM, Peh SQ, Sia YY, Li G, Mulawadi FH, et al. ChIA-PET analysis of transcriptional chromatin interactions. Methods 2012;58:289–299. PMID: 22926262.


Fig. 1
Perspectives of epigenome research. MeDIP, methylated DNA immunoprecipitation-sequencing; MIRAseq, methylated CpG island recovery assay-sequencing; iPS, induced pluoripetent stem cell; mC-seq, methylated cytosine-sequencing; CHIA-PET, chromatin interaction analysis using paired end tags; T2D, type 2 diabetes; GWAS, genome-wide association study; ENCODE, Encyclopedia of DNA Element; EWAS, epigenomewide association study; hmC-seq, hydroxyl methyl cytosine-sequencing; IDH, isocitrate dehydrogenase; IHEC, International Human Epigenome Consortium.
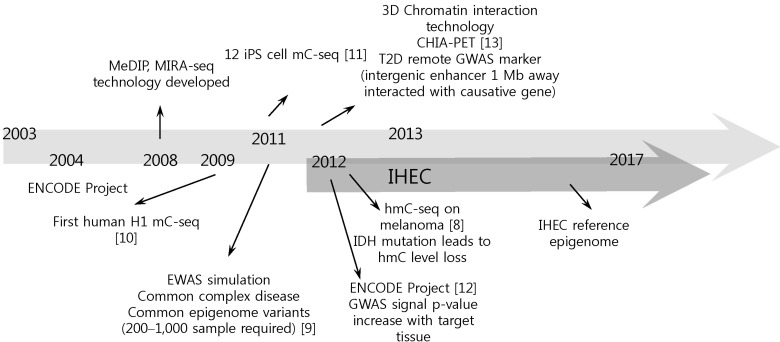
Table 1.
Major international efforts after the Human Genome Project (HGP) completion in 2003
Table 2.
1,000 Epigenome production scheme
iPS, induced pluoripetent stem cell; IEO, Istituto Europeo di Oncologia; IFOM, Istituto FIRC di Oncologia Molecolare; IIT, Istituto Italiano di Technologia; DEEP, Deutsches Epigenom Programm; GI, gestrointestinal; FACS, fluorescent activated cell sorting; PP, pancreatic polypeptide producing; HIV, human immunodeficiency virus; HPV, human papillomavirus; GU, genitourinary.
Table 3.
Definition of International Human Epigenome Consortium (IHEC) epigenome set
- TOOLS
-
METRICS

-
- 34 Crossref
- 8,229 View
- 108 Download
- Related articles in GNI





