 |
 |
- Search
| Genomics Inform > Volume 21(3); 2023 > Article |
|
Abstract
Mycobacterium tuberculosis (Mtb) is the causative agent of tuberculosis, one of the most deadly infections in humans. The emergence of multidrug-resistant and extensively drug-resistant Mtb strains presents a global challenge. Mtb has shown resistance to many frontline antibiotics, including rifampicin, kanamycin, isoniazid, and capreomycin. The only licensed vaccine, Bacille Calmette-Guerin, does not efficiently protect against adult pulmonary tuberculosis. Therefore, it is urgently necessary to develop new vaccines to prevent infections caused by these strains. We used a subtractive proteomics approach on 23 virulent Mtb strains and identified a conserved membrane protein (MmpL4, NP_214964.1) as both a potential drug target and vaccine candidate. MmpL4 is a non-homologous essential protein in the host and is involved in the pathogen-specific pathway. Furthermore, MmpL4 shows no homology with anti-targets and has limited homology to human gut microflora, potentially reducing the likelihood of adverse effects and cross-reactivity if therapeutics specific to this protein are developed. Subsequently, we constructed a highly soluble, safe, antigenic, and stable multi-subunit vaccine from the MmpL4 protein using immunoinformatics. Molecular dynamics simulations revealed the stability of the vaccine-bound Toll-like receptor-4 complex on a nanosecond scale, and immune simulations indicated strong primary and secondary immune responses in the host. Therefore, our study identifies a new target that could expedite the design of effective therapeutics, and the designed vaccine should be validated. Future directions include an extensive molecular interaction analysis, in silico cloning, wet-lab experiments, and evaluation and comparison of the designed candidate as both a DNA vaccine and protein vaccine.
Tuberculosis, an infectious disease caused by the pathogen Mycobacterium tuberculosis (Mtb), ranks as the 10th leading cause of death worldwide. The World Health Organization (WHO) reported approximately 1.6 million and 1.4 million deaths in 2018 and 2019, respectively [1,2]. In 2019, two-thirds of global cases were reported in eight countries, with India accounting for the highest percentage (26%), and its neighboring country, Bangladesh, ranking seventh [1]. Tuberculosis has become a significant challenge to medical science due to the diminishing effectiveness of older broad-spectrum antibiotics and instances of treatment failure. These factors have contributed to the development of antituberculosis drug resistance and the spread of multi-drug resistant (MDR) and extensively drug-resistant (XDR) Mtb strains [1-4]. MDR Mtb is characterized by resistance to at least two first-line antibiotics, while XDR Mtb is resistant to at least one quinolone and one of three second-line injectable agents (amikacin, kanamycin, or capreomycin) [5,6]. In 2019, an estimated 78% of MDR tuberculosis was reported among nearly half a million rifampicin-resistant tuberculosis patients [1]. In most cases, effective injectable agents could be administered [7]. Mtb can exhibit either primary (transmitted) resistance or secondary (acquired) resistance [6,8]. Several mechanisms of drug resistance have been studied, and approximately 74 resistance-related genes have been identified [9]. Examples of mutant/resistance-conferring genes include rpoB and ponA1 for rifamycins; katG, and inhA for isoniazid; gyrA and gyrB for levofloxacin and moxifloxacin; Rrs for amikacin, capreomycin, and kanamycin; Eis for kanamycin; and tlyA for capreomycin [10,11].
Vaccines minimize the misuse of antibiotics, which in turn helps prevent the spread of antibiotic-resistant bacterial strains [12]. However, broad-spectrum antibiotics may interact with the human microbiome, while vaccines tend to minimally disrupt the microbiota [13]. The bacillus Calmette-Guerin (BCG) vaccine, which is the only licensed vaccine against tuberculosis, exhibits a protective effect for only 10-15 years and its protection gradually wanes with age in adults. As a result, while the BCG vaccine is effective in providing protection against miliary tuberculosis in childhood, its efficacy in protecting against adult pulmonary tuberculosis varies widely, ranging from 0% to 80% [14-17]. Differences in the BCG strain [18], the genetic backgrounds in diverse populations [19], geography [20], and immunization strategies [21] all play crucial roles in the varying efficacy of BCG vaccines. Furthermore, complications associated with the BCG vaccine are more common in immunocompromised patients [22-24]. This failure contributes to the global disease burden of adult pulmonary tuberculosis. Currently, 14 vaccine candidates are undergoing clinical trials [25-27], and several vaccine development studies are targeting 48 Mtb genes (http://www.violinet.org/). Despite these efforts, scientists are still struggling to achieve satisfactory results. Therefore, the development of safe and effective vaccines is urgently needed to address the antibiotic resistance crisis and combat MDR and XDR Mtb strains.
Advances in bioinformatics tools have expedited the process of vaccine development. These computational tools are regularly used in various methods to analyze the published genomes and proteomes of pathogens, with the aim of identifying new vaccine candidates [28,29]. We have used subtractive proteomics to identify potential therapeutic targets against Brucella abortus strain 2308 [30] and Bartonella bacilliformis [31]. Moreover, we have employed an immunoinformatics approach to design vaccine candidates against severe acute respiratory syndrome coronavirus 2 [32] and West Nile virus [33]. The detailed protocols of vaccine development through the immunoinformatics approach are well established [34-37]. Pre-clinical studies (both in vivo and in vitro) of the vaccines developed through the immunoinformatics approach against Chlamydia trachomatis [38,39], Streptococcus agalactiae [40], Rickettsia prowazekii [41], viruses (Epstein–Barr virus [42], Crimean-Congo hemorrhagic fever virus [43]), and protozoan parasite (Toxoplasma gondii [44]) have proven the efficacy of the resulting vaccines.
In this study, we used subtractive proteomics with various parameters to identify new vaccine targets from the reference strain M. tuberculosis H37Rv, and performed a conservancy analysis in 22 other virulent MDR and XDR Mtb strains. Given that more vaccines have been licensed in the last 40 years than new antibiotics [13], we emphasized vaccine development for short-listed targets. We augmented our study by using the immunoinformatics approach to design an immunogenic recombinant subunit vaccine against Mtb strains, which would stimulate both innate and adaptive immune responses [45-48]. Our results identified the MmpL4 (mycobacterial membrane protein Large) protein as a potential therapeutic target. Mtb contains 13 MmpL proteins that export bulky, hydrophobic substrates to support the ability to infect and persist in the host [49, 50]. Among these 13 MmpL proteins, MmpL4 plays a role in the transport of iron/siderophore (carboxymycobactin). This siderophore exportation and iron importation are facilitated by the MmpL4 protein, which interacts with MmpL5 and the periplasmic adaptor proteins MmpS4/5 (mycobacterial membrane protein-small) [51]. Iron is an essential nutrient for many cellular processes, and Mtb produces siderophores to capture iron from hosts. Disruption of proper carboxymycobactin exportation has a detrimental effect on Mtb survival and growth in vivo and in vitro [52]. MmpL4 is a virulence-conferring protein essential for maintaining normal growth in the murine tuberculosis model. Mutational disruption in MmpL4 causes impaired growth of the Mtb strain and renders it avirulent [53]. Although the MmpL4 protein is part of the RND (resistance, nodulation, and cell division proteins) family, it does not contribute to intrinsic drug resistance or antituberculosis drug resistance, making the MmpL4 protein a promising antituberculosis drug target [52,53]. Furthermore, iron restriction is a common strategy to combat pathogenic infections [49]. Therapeutics designed to disrupt or block the interaction of the MmpL4 protein with MmpL5 and MmpS4/5 proteins could lead to iron deficiency and subsequent eradication of Mtb infection in the host. Given that Mtb has already developed resistance against many antibiotics and evolved into MDR and XDR Mtb, targeting a protein that has not been exposed to any therapeutics would be an ideal candidate to treat MDR and XDR Mtb cases. Currently, there is no drug or antibiotic available that targets the MmpL4 protein. Considering these issues, we believe that MmpL4 would be a suitable candidate for designing therapeutics to combat MDR and XDR cases. We designed an epitope-based subunit vaccine against the MmpL4 protein to combat antimicrobial resistance. However, the designed vaccine requires further wet-lab evaluation and validation to prove its efficacy in vivo and in vitro. Our future goals for this study include the wet-lab expression and evaluation of the designed vaccine as a DNA vaccine and protein vaccine [39,43].
The subtractive genomic approach was employed to identify novel drug targets and vaccine candidates for the entire proteome of M. tuberculosis H37Rv (a representative strain of Mtb). The overall workflow is illustrated in Fig. 1.
We retrieved the entire proteome of M. tuberculosis H37Rv (Assembly GCA_000195955.2) [54] from the NCBI Genome database. We selected this particular genome since it is recognized as a reference proteome for Mtb.
The essential proteins of M. tuberculosis H37Rv were predicted through the Database of Essential Gene (DEG) server v15.2 [58], which contains 53,885 essential genes and 786 essential non-coding sequences. This server is broadly used in the subtractive genomics/proteomics approach to identify essential proteins in organisms [59-61]. The protein hit settings of an expectation value (E-value) ≤ 10-100, identity ≥ 25%, and bit score > 100 were used to filter essential proteins for this pathogen [57,62].
A comparison was conducted between the host and pathogen to identify pathogen-specific pathways (herein termed “unique pathways”) by using the three-letter organism codes “has” and “mtu” for Homo sapiens (host) and M. tuberculosis H37Rv, respectively, through the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Database [66].
The identified host non-homologous proteins of M. tuberculosis H37Rv were subjected to BLASTp through the KAAS server [67] in the KEGG database. Proteins that were involved in unique pathways were identified as unique proteins. Those proteins that were assigned KO (KEGG Orthology) but did not participate in common host-pathogen pathways were also classified as unique proteins. Proteins that were involved in common pathways, as well as those without KO assignments, were excluded from the selection. Hypothetical unique proteins were also omitted from this study.
M. tuberculosis H37Rv is a Gram-positive bacterium. As such, we can predict the subcellular localization of previously selected unique proteins—which were non-homologous to the host and specific to the pathogen, and essential—to be extracellular, cell wall, membrane, or cytoplasmic. To ensure the highest accuracy in our predictions, we utilized CELLO v.2.5 [68], PSORTb v3.0.2 [69] and PSLPred [70]. If a particular protein's location was predicted to be the same by either all three or any two of these servers, we adopted that location for our druggability analysis. However, if each server predicted a different location for a specific protein, we excluded that protein from our study.
Unique proteins were screened using the DrugBank v5.1.4 database [71] to assess their novelty as drug targets. The DrugBank Database includes numerous FDA-approved drugs and displays protein targets along with their respective drug IDs. A BLASTp analysis was conducted using default parameters, and proteins that did not match any drug were chosen for further investigation as potential novel drug targets for M. tuberculosis H37Rv [30]. Additionally, the molecular weight of these unique proteins was calculated using the ExPASy/ProtParam server [72].
A list of 83 organisms found in the gut flora of healthy humans was compiled from the existing literature (Supplementary File 1) [73]. An NCBI BLASTp analysis was conducted on these unique proteins, with parameters set to an E-value < 0.001, sequence similarity of < 35%, and < 10 hits. The results were then cross-referenced with another set of gut microflora data [74] that contained 95 organisms (Supplementary File 2). Unique proteins that showed <10 hits and <35% similarity were selected for further analysis.
Finally, unique proteins that had a precise location, were predicted as novel drug targets, had a molecular weight <110 kDa, and exhibited low similarity with human gut microflora were selected for an analysis of human anti-targets.
The term "anti-target" is used to refer to essential human proteins. In total, 210 anti-targets reported in the literature with their accession numbers (Supplementary File 3) [73] were obtained from the NCBI Protein Database. Previously selected novel drug targets were subjected to BLASTp in NCBI analysis, setting the parameters as an E-value ≤10–4, identity <25%, and query length >30%. Only membrane proteins were selected for antigenicity analysis.
To determine whether the selected proteins were preserved in other virulent Mtb strains, we subjected both cytoplasmic and membrane vaccine targets to BLASTp in NCBI, comparing them against 22 virulent Mtb strains documented in the literature [77,78]. The following cut-off parameters were used to screen proteins: an E-value <0.0001, query cover >80%, and similarity >90%. We selected proteins that passed the screening for protein-protein interaction (PPI) analysis.
Proteins form complex interaction networks to function properly. The analysis of these PPIs aids in understanding biological pathways and the structural organization of cells [79]. The most promising metabolic functional associations were identified for selected highly conserved proteins through these interactions. Using STRING v11.0 [80], PPIs for selected proteins were generated with high confidence (>0.700). Additionally, the accession number, gene name, and functions of these interacting proteins were curated from the UniProt database [81].
A pool of 9-mer cytotoxic T lymphocyte (CTL; CD8+) epitopes was created using the NetCTL1.2 server [83], with restrictions for 12 human MHC class I supertypes and default parameters. A pool of 15-mer helper T lymphocyte (HTL; CD4+) epitopes was generated using the IEDB MHC II server [84]. The HTL epitopes were chosen based on mouse alleles with a percentile rank of less than 10 and an IC50 value of less than 500 nM.
All short-listed T-cell and B-cell epitopes were screened using the VaxiJen v 2.0 server to check their antigenicity score. The TMHMM v.20 server was used to predict topology, whereas the AllerTop server [87] and ToxinPred server [88] were used to check the allergenicity and toxicity of these epitopes, respectively. Additionally, the immunogenicity of CTL epitopes was evaluated through the IEDB MHC-I Immunogenicity server [36], and the IFNepitope sever [89] was used to predict interferon-γ (IFN-γ) inducing HTL epitopes.
Short-listed T-cell epitopes were analyzed using the IEDB Population Coverage tool [90] to predict worldwide coverage. Subsequently, the HLA alleles and their corresponding T-cell epitopes were uploaded to the server.
Potential CTL, HTL, and BCL epitopes were combined to construct a subunit vaccine. An AAY linker was utilized to separate CTL epitopes, while the GPGPG linker and KK linker were employed to attach the HTL and BCL epitopes, respectively [91]. In addition, an EAAAK linker separated the L7/L12 Ribosomal protein, a Toll-like receptor-4 (TLR-4) agonist [92], from the universal PADRE sequence at the N-terminal end of the vaccine [93]. An EGGE linker joined the invasin peptide with another PADRE sequence at the C-terminal end of the vaccine [94].
The ExPASy-ProtParam server was used to predict several physiochemical properties of the vaccine construct, including molecular weight, theoretical PI, instability index (II), half-life, and aliphatic index (AI). Furthermore, the VaxiJen server was utilized to predict the antigenicity of the constructed vaccine. The AlgPred server [95] and SOLpro tool [96] were employed to predict the allergenicity and protein solubility for the Escherichia coli expression system.
The Robetta server [96] was used to predict the three-dimensional (3D) structure of the constructed vaccine. This structure was then refined using the GalaxyRefine server [97]. Next, only the best-refined model was evaluated through the PROCHECK [98] and Verify3D [99] algorithms on the SAVES server and compared with the initially modeled structure. The ProSA webserver [100] was used to predict the Z-score. Finally, a refined structure was selected for docking with human TLR-4.
The PDB structure of human TLR-4 (PDB id: 4G8A) was prepared using the SWISSPDB viewer for docking purposes. The ClusPro 2.0 server [101] was utilized to perform molecular docking of the refined vaccine protein with the TLR-4 monomer. The docked complex with the lowest binding energy was then subjected to molecular dynamics (MD) simulation in YASARA [102], utilizing the AMBER14 force field. Initially, the system was cleaned and optimized. The TIP3P model was implemented, with Na/Cl ions added at a density of 1.012 g/cm3. The particle mesh Ewald method was used to calculate long-range electrostatic interactions at a distance of 8 Å. The physiological system of the complex was set at 298K with a pH of 7.0 and 0.9% NaCl [103]. A cubic simulation cell was created (126.4688 Å × 126.4688 Å × 126.4688 Å), and the periodic boundary condition was maintained. The MD simulation was then run with a time step of 2.50 fs [103]. Finally, the root mean square deviation (RMSD), root mean square fluctuation (RMSF), the radius of gyration (Rg), and solvent accessible surface area (SASA) were calculated from the trajectory. Non-bonding interaction analysis was carried out using the PDBsum server [104], which reports the number of non-bonded interactions, hydrogen bonds, salt bridges, disulfide bonds, and their respective contact residues. An immune simulation analysis was conducted using the C-ImmSim server to evaluate the primary and secondary immune responses to the designed vaccine [104]. The doses were administered at intervals of 1, 84, and 168 time steps, with each time step equivalent to 8 hours [105].
The reference strain M. tuberculosis H37Rv, retrieved from the NCBI Genome Browser, contains 3906 proteins. These proteins were short-listed through screening with different web-based bioinformatics servers, and the results are shown in Table 1.
The CD-Hit suite identified 35 paralogous clusters (with a similarity greater than 80%) within the entire proteome of the pathogen. It also detected a total of 46 redundant or duplicate sequences, which could potentially lead to false-positive results in subsequent analyses. Consequently, these duplicate sequences were omitted from further examination [30]. Proteins larger than 100 amino acids are more likely to play a role in the essential metabolic pathways of the organism than smaller proteins (those less than 100 amino acids in length). Therefore, even though they are unique to the organism, a total of 292 protein sequences were excluded on the assumption that they were unlikely to be essential proteins [62].
The most critical criterion for a therapeutic target is its potential indispensability for the pathogen's survival [106]. Out of 3565 non-paralogous sequences of the pathogen, only 580 proteins met the screening criteria (E-value ≤10-100, identity ≥25%, bit score >100) through the DEG v15.2 server. Therefore, these 580 proteins were ultimately identified as essential proteins.
Therapeutics targeting proteins from the pathogen that are homologous to those in the host (H. sapiens) could potentially disrupt host metabolism and trigger cytotoxic reactions. However, out of the total, 250 essential proteins demonstrated hits above the threshold (E-value >10-4) with the RefSeq proteome of humans. Consequently, these proteins, believed to have host homologs, were omitted from our analysis. This exclusion reduces the likelihood of cross-reactivity when a therapeutic is administered to fight the pathogen.
The metabolic pathways of M. tuberculosis H37Rv were analyzed in comparison to human metabolic pathways using the KEGG server. Both the host (“has”) and the pathogen (“mtu”) metabolic pathways were curated from the KEGG Pathway Database. This comparison was conducted to identify shared pathways and those specific to the pathogen. Essential proteins from the host that were non-homologous were subjected to BLASTp analysis via the KAAS server at KEGG. Out of 330 proteins, 117 were found to be involved in shared pathways. A total of 166 proteins were not assigned a KO, suggesting that they are not metabolic proteins. Consequently, these 28 proteins were also excluded from our study. The remaining 47 proteins were assigned to the KO, indicating their involvement in pathogen-specific pathways. However, six proteins were hypothetical and therefore excluded from the study (Supplementary Table 1). As a result, 41 proteins associated with pathogen-specific pathways were short-listed for further investigation [73,107].
Advances in bioinformatics, cheminformatics, and genome sequencing, when combined with experimental data, have shown that several key factors contribute to the identification of potential drug targets in pathogenic organisms. These factors include the prediction of subcellular localization, the novelty of the drug target, molecular mass (kDa), and the presence of transmembrane helices [108-110].
Cytoplasmic proteins serve as suitable drug targets, while membrane proteins are viewed as potential drug targets and vaccine candidates [59]. A short list of 41 proteins from the Gram-positive bacterium M. tuberculosis H37Rv was subjected to CELLO, PSORTb, and PSLPred to predict subcellular localization with improved accuracy. Out of the 41 proteins, 23 were predicted to be cytoplasmic proteins, 16 were predicted to be membrane proteins, and one was predicted to be an extracellular protein. However, all three servers were unable to accurately predict the location of the remaining protein.
The primary objective of this study was to identify new vaccine targets for MDR and XDR Mtb strains. To this end, we screened 41 pre-selected proteins through the DrugBank Database to determine their potential novelty as drug targets. Of these 41 proteins, 11 demonstrated significant homology with drug targets of other pathogenic strains. Despite the administration of antibiotics to eliminate other pathogenic bacteria, there is a possibility that these 11 proteins may be exposed to antibiotics or antibacterial drugs. Consequently, the remaining 30 proteins, which did not match any known drug targets, were deemed to be novel drug targets.
The 41 short-listed proteins were screened using NCBI BLASTp to minimize the risk of inadvertently inhibiting the gut microflora present in a healthy human. These gut prokaryotic symbionts play a crucial role in assimilating poorly digestible dietary components, degrading xenobiotics, synthesizing vitamins, and providing resistance against colonization by opportunistic bacteria and other pathogens [111-114]. Consequently, any deterioration of these gut microflora strains could result in nutritional deficiencies in the host and a weakened first-line defense against pathogen invasion [112]. Initially, the 41 short-listed proteins were compared with 83 organisms cited in the literature, using parameters set at an E-value of <0.001, sequence similarity of <35%, and <10 hits. The results were then cross-verified with a set of 95 organisms mentioned in other literature. Ultimately, only nine proteins successfully passed the gut microflora non-homology screening.
It is feasible to experimentally study proteins with a molecular mass of less than 110 kDa. However, proteins with a molecular mass greater than 110 kDa are not suitable as therapeutic targets due to their larger structures [59,109]. Therefore, we determined the molecular weight of the 41 short-listed unique proteins using the ExPASy/ProtParam server.
Finally, five membrane proteins were chosen based on their minimal similarity to human gut microflora, the novelty of their drug target, and a molecular weight of less than 110 kDa. The screening data can be found in Table 2.
The removal of unique proteins that are homologous to human anti-targets is crucial in the identification of an appropriate therapeutic target. Human 'anti-targets' encompass the ether-a-go-go related gene (hERG), the constitutive androstane receptor, the pregnane X receptor, P-glycoprotein, and certain membrane receptors such as muscarinic M1, adrenergic a1a, serotonergic 5-HT2C, and dopaminergic D2 [115]. Between 1960 and 1999, several drugs were withdrawn due to adverse reactions with human anti-targets [116], such as benoxaprofen in 1982 [117] and trovafloxacin in 1999 [118]. Therefore, the five selected proteins were subjected to BLASTp on the NCBI server, with parameters set to an E-value ≤10±, identity <25%, and query length >30%. Fortunately, all five proteins successfully passed the screening, thereby reducing the potential for adverse side effects when therapeutics are administered to humans.
Reverse vaccinology has been demonstrated to be a practical approach for discovering vaccine candidates [119]. Emerging safe recombinant vaccines, based on antigenic protein sequences, are becoming the most appealing and cost-effective solution in the battle against infectious diseases. An ideal antigenic protein should have an antigenicity score greater than 0.4, as determined by the VaxiJen server, and should possess transmembrane helices [120]. Of the five proteins examined, all had transmembrane helices, but only one protein, NP_214964.1 (transmembrane transport protein MmpL4), exhibited antigenicity with a VaxiJen score exceeding 0.4 (Supplementary Table 2). Consequently, this MmpL4 protein (‘NP_214964.1’) has been selected as a potential vaccine candidate, in addition to being considered a novel drug target.
A list of 22 other virulent MDR and XDR Mtb strains has been reported in the literature (Supplementary Table 3) [77,78]. One membrane vaccine candidate (transmembrane transport protein MmpL4, NP_214964.1) of M. tuberculosis H37Rv was subjected to BLASTp on the NCBI server (E-value <0.0001, query coverage >80%, sequence similarity >90%) against these 22 MDR and XDR Mtb strains. This protein was found to be present in all 22 virulent strains with at least 99% similarity. Therefore, infections caused by these 23 strains in humans could potentially be treated if therapeutics are designed against any of these essential, unique proteins of Mtb.
PPIs, which are essential for proper functioning, take place in the cellular network, biological pathways, and structural organization of a cell. One of the primary challenges in bioinformatics is the computational analysis of these intricate networks, which aids in understanding whole-cell engineering, drug targets, and cellular activities [79]. The STRING database, with a high confidence score of 0.700, indicates that NP_214964.1 interacts with seven proteins (Fig. 2). Any disruption of these interactions is expected to affect several vital pathways of the pathogen.
NetCTL predicted a total of 245 epitopes, which are restricted for MHC class I human super types such as A1, A2, A3, A24, A26, B7, B8, B27, B39, B44, B58, B62. Out of these, 37 epitopes were found to be antigenic, immunogenic, non-toxic, and non-allergic, with an external topology. These characteristics are crucial for CTL epitope selection [125]. Ultimately, four peptide regions containing epitope clusters (23MIHAFAVPIILGW33, 221SIITVVLLLITVGVEL236, 380VVVRWPLPVLV360, and 514SVLLWQHILAIHLHWL529) were selected for vaccine construction (Supplementary Table 4).
In contrast to CTL, the IEDB MHC II server predicted 50 epitopes restricted for mouse H-2-I alleles. Of these, 18 epitopes were predicted to be antigenic, non-toxic, and non-allergic with an external topology. Only 12 HTL epitopes were found to be capable of inducing IFN-γ secretion (Supplementary Table 5). Finally, three peptide regions containing epitope clusters (377VGTVVVRWPLPVLVA391, 878NAGLVFAVTMASMAVSDL895, and 925AALLGRWFWWPLRVRSRP943) were selected for vaccine construction.
A total of 81 BCL epitopes (20-mer) were predicted using the ABCpred server. However, only 11 of these epitopes demonstrated antigenicity, non-toxicity, and non-allergenicity. These selected epitopes were then cross-verified using the IEDB BepiPred 2.0 method. Ultimately, five peptide regions containing epitope clusters (49LEAVGQERSVSLSPKDAPSF68, 181YVTGPSALAADMHHSGDRSM200, 481RPEGTTMDHTSIPFQISMQNAGQLQTIKYQ520, 531LTRMHSLMAEMASTTHRMVGDTEEMKE557, and 932FWWPLRVRSRPARTPTVPSE951) epitopes were selected for vaccine construction (Supplementary Table 6).
Population coverage was determined for both CD8+ T cells and CD4+ T cells, along with their respective HLA alleles. The highest coverage for CTL was reported in North America and the United States (100%), while the highest for HTL was found in Germany (93%). Therefore, approximately 98.84% of the global population could be covered through the selected CTL epitopes, and around 86.76% through the selected HTL epitopes. These results are shown in Fig. 3.
A vaccine composed of 443 amino acids was engineered by combining selected CTL, HTL, and BCL epitopes. The 50s ribosomal protein L7/L12 from Mtb (130 amino acids) was attached at the N-terminal end to serve as an adjuvant, stimulating TLR-4. An empirical α-helical linker (EAAAK) was employed to connect the adjuvant with the PADRE (AKFVAAWTLKAAA) sequence. The AAY, GPGPG, and KK linkers were respectively inserted between the CTL, HTL, and LBL. The EGGE linker was utilized to connect the invasin peptide (TAKSKKFPSYTATYQF) with the LBL epitope at the C-terminal end (Fig. 4A).
The physiochemical properties of the constructed vaccine indicate that it is highly stable (II 36.14), with an estimated half-life of 30 hours in mammalian reticulocytes (in vitro). The vaccine is also expected to be thermostable (AI 93.70). Moreover, the vaccine protein is safe, antigenic, and likely soluble in the E. coli expression system. The results for all parameters are detailed in Table 3. Three tertiary structures were created using the Robetta server and the ab initio modeling algorithm, as no suitable template was available for homology modeling. Out of five models, model 3 had 87.5% of residues in the most favored region. The GalaxyRefine server refined model 3, generating five models, with the first one (Fig. 4B) being the best refined. Furthermore, the PROCHECK server demonstrated that the refined model was an improvement over the initial model 3, with 88.8% of residues in the most favored region (Fig. 4C). The refined model deviated from its initial design by an RMSD of 0.312 Å. The Verify3D and Z-score of the refined model were 89.03 and –6.33, respectively. The parameters of the refined models can be found in Supplementary Table 7.
The ClusPro 2.0 server generated 30 complexes of TLR-4 bound to the vaccine. Among these, cluster 04 emerged as the best-docked complex, exhibiting the lowest binding energy (-1,114.00 J/Mol). Numerous studies have highlighted the roles of TLR-4 in immunosurveillance and the eradication of the intracellular bacterial pathogen Mtb [126-129]. TLR4 is expressed on both cytoplasmic and endosome membranes and signals through TRIF- and MYD88-dependent pathways [130,131], thereby facilitating immunosurveillance against tuberculosis [132]. To assess the stability of the TLR-4 complex bound to the vaccine, a 20 ns molecular dynamic simulation was conducted. As can be seen in Fig. 5A, the RMSD values increased during the initial phase of the simulation. However, after 10 ns, the RMSD peak appeared to stabilize, maintaining its integrity throughout the 20 ns simulation time. The RMSD values of the systems remained well below 2.5Ǻ for the entire simulation duration. The RMSD of the protein or protein complex signifies the overall stable nature of the protein. Fig. 5B illustrates the rigid nature of the epitope and protein complex [133].
The RMSF also provides insight into the flexible regions of the protein [134]. Fig. 5C illustrates the consistent nature of the epitope and protein complex. No excessive fluctuations were noted, although certain amino acids such as Gly363, Lys560, Gln562, Lys615, and Thr626 exhibited higher RMSF compared to other compounds. Conversely, the degree of compactness can be verified by the Rg. As seen in Fig. 5D, the Rg profile of the vaccine and TLR-4 complex initially decreased slightly after 5 ns before reaching a steady state. The overall peak of the Rg did not exhibit significant fluctuations, indicating a rigid state. The SASA value of the epitope and protein complex was initially lower, but it increased beyond the initial phase. The lower SASA value accounts for the protein's contracted nature [134], and the overall SASA peak signifies the robust nature of the complex. These data collectively suggest the stability of the TLR-4 vaccine complex, which could facilitate the transportation of the vaccine within the antigen-presenting cell. The PDBsum server reported four hydrogen bonds (A-C: 31Glu-249Thr, 32Val-211His, 35Asn-73Lys, 605Glu-126Thr) and 78 non-bonded interactions between TLR-4 (Chain A) and the vaccine (chain C). In contrast, two hydrogen bonds (B-C: 39Lys-219Pro, 143Glu-228Trp), one salt-bridge (B-C: 136Glu-227Arg), and 30 non-bonded interactions were reported between the MD molecule (Chain B) and the vaccine (chain C). All six hydrogen bonds were between 2-3 Å, indicating strong interactions between the vaccine-bound TLR-4/MD complex [135]. The inclusion of a salt-bridge interaction could enhance the biomolecular stability of the vaccine-TLR complex [136-138]. Hydrogen bond and salt-bridge interactions are shown in Table 4 and Fig. 6.
Immune stimulation of the multi-epitope vaccine was conducted to determine if the epitopes and adjuvants could generate sufficient adaptive immunity. The analysis clearly demonstrated that the designed vaccine has the potential to trigger a typical immune response. A simulation involving three doses showed a subsequent increase in the primary immune response and stimulation of the secondary immune response. The vaccine was projected to significantly induce primary immune responses incrementally after each of the three injections (Fig. 7). Moreover, the secondary immune response was stimulated, and the primary immune response gradually increased after each dose (Fig. 7A). The number of B cells (Fig. 7B and 7C), plasma B cells (Fig. 7D), helper T cells (Fig. 7E and 7F), cytotoxic T cells (Fig. 7G and 7H) and antigen-presenting cells such as dendritic cells and macrophages (Fig. 7I and 7J) increased significantly after administration.
Tuberculosis, the leading cause of death from a single infectious agent, continues to confound thousands of scientists in this modern era [2]. The global prevalence of MDR and XDR Mtb strains can be attributed to the transmission of gene resistance through horizontal gene transfer and the acquisition of gene resistance via vertical gene transfer. The investigation of non-resistant gene products is crucial for the development of new antimicrobial targets and vaccine candidates. Vaccines have the potential to stimulate immune responses for decades without inducing resistance [139]. The human gut microflora is less likely to be impacted by vaccines compared to broad-spectrum antibiotics, which target multiple bacterial species simultaneously [140]. Traditional vaccine development is a time-consuming, laborious, and expensive process [35,141]. Several experimental studies have highlighted the immunoinformatics approach as a promising strategy for designing highly efficient, immunogenic, and safe vaccines [35].
Several drug repositioning studies have been conducted on druggable proteins of Mtb, in which multiple drugs were available either for selected proteins of Mtb or homologous proteins of other pathogenic species [78,121]. Two groups utilized subtractive proteomics to screen out promising therapeutic compounds [122,123], and two other groups screened out vaccine targets [77,124]. However, neither group conducted immunoinformatics to design a vaccine, nor did they predict MmpL4 as a potential drug target or vaccine candidate. One group carried out an immunoinformatics study, selecting 38 highly expressed Mtb proteins in vivo (in both humans and mice), but the study was limited to screening only B-cell and T-cell epitopes [142]. Although two other groups used immunoinformatics to design vaccines against Mtb, they did not use a subtractive proteomics approach to identify pathogen-specific, essential targets that are less homologous to gut microflora [9,143]. Several crucial steps of subtractive proteomics include host non-homology analysis, essential protein analysis, pathogen-specific pathway analysis, non-resistance analysis, subcellular localization analysis, and gut microbiome analysis.
In our study, we utilized a computational proteome subtraction approach to identify non-resistant, essential, antigenic membrane proteins. We predicted the transmembrane transport protein MmpL4 (NP_214964.1) as the most promising candidate for vaccine development. The MmpL4 protein was also predicted to be non-homologous to human anti-targets and showed limited similarity to gut microflora (less than 28% similarity with only five hits). The MmpL4 protein plays a crucial role in iron transportation. Mtb sustains its growth within the host by seizing and importing iron, a vital nutrient, into its cytoplasm. When cellular iron is scarce, the pathogen synthesizes siderophores (mycobactins and carboxymycobactins) to capture iron, thereby evading the host's immune system [51]. The MmpL4 protein, in conjunction with the MmpL5 protein, synthesizes and transports siderophores. The siderophore export accessory proteins MmpS4 and MmpS5, which are anchored in the membrane, facilitate this transportation. MmpL4 has been shown to interact with virulence factors (MmpS4 and MmpS5) (Fig. 2) [51]. Therefore, disrupting the function of the transmembrane transport protein MmpL4 and its interactions with other transporter proteins could result in iron deficiency in the pathogen, ultimately leading to the pathogen's death. The DrugBank Database results revealed no existing drug or homologous protein for MmpL4 (NP_214964.1), indicating the novelty of MmpL4 as a species-specific drug target. This protein is also predicted to be a potential antigenic protein and was found to be 99% conserved in 22 MDR and XRD strains of Mtb. Subsequently, we employed an immunoinformatics approach on the MmpL4 protein to design an efficient multi-epitope vaccine candidate. Therefore, our findings in this study have contributed advanced knowledge compared to previous studies
Antigenic epitopes, which include T-cell (CD8+ and CD4+) and B-cell epitopes, are crucial for designing an ideal multi-epitope vaccine and generating a specific immune response against predicted antigens [37]. The roles of CD8+ T cells and CD4+ T cells in responses against Mtb have been extensively reviewed by Prezzemolo et al. (2014) [144] and Lin et al. (2015) [145]. In short, it is well established that CD8+ T cells express perforin, granzyme, and granulysin, which induce apoptosis in Mtb-infected cells, such as macrophages, in both human and mouse models during antigen-specific responses [145-148]. Numerous studies have identified CD4+ T cells as crucial for controlling tuberculosis infections [149-151]. IFN-γ plays an important role in phagocytosis and oxidative bursts in Mycobacterium spp. [152]. Moreover, the activation of IFN-γ and tumor necrosis factor-α, mediated by Mtb-specific CD4+ T cells, recruits monocytes and granulocytes, thereby enhancing their antimicrobial activities [153-156]. CD4+ T cell-induced IFN-γ stimulates macrophages to synthesize nitric oxide, leading to the clearance of this pathogen [157-160]. Therefore, both CD8+ and CD4+ T-cell epitopes were screened from the MmpL4 protein, and the selection process was based on several critical properties such as antigenicity, immunogenicity, non-toxicity, non-allergenicity, external topology, and cytokine production [104]. Four CD8+ T-cell peptides and three CD4+ T-cell peptides containing multiple epitopes (clusters) were selected for the subunit vaccine design. Conversely, both naïve and memory B cells are found in the lungs of humans infected with tuberculosis [48], and they assist in antigen presentation to T cells, cytokine production, and Mtb-specific antibody generation [48]. Rao et al. (2015) [161] highlighted the role of B cells in the antibody-mediated response against tuberculosis. Kringelum et al. (2013) [162] reported that LBL epitopes are more stable than discontinuous epitopes, which is why we only screened LBL epitopes in our study. Five peptide regions containing LBL epitope clusters were selected for the vaccine design. The vaccine was assembled with adjuvants and linkers. The adjuvant 50s ribosomal protein L7/L12 of Mtb (130 amino acids), which has proven effective in stimulating TLR-4 [92], was used in the vaccine design, while the EAAAK linker enhances the stability and bi-functional catalytic activity of the fusion protein [163]. The PADRE sequence (AKFVAAWTLKAAA) was incorporated to address issues caused by highly polymorphic HLA alleles [164]. The invasin peptide (TAKSKKFPSYTATYQF) was used to enhance the immune response by the adenoviral DNA vaccine [165].
The designed vaccine was antigenic, non-toxic, highly stable, hydrophilic, soluble, and found to be stable as a vaccine-TLR4/MD docked complex through molecular dynamics simulation. Our group has computationally designed an adenoviral DNA vaccine for West Nile virus [33]. Adenoviral vaccines can be produced at a relatively low cost and at higher tiers of vaccines, making this platform widely studied and extensively evaluated for vaccine development [166-170]. Pharmaceutical companies such as Johnson & Johnson, AstraZeneca, CanSino Biologics Inc., and the Gamaleya Research Institute have recently marketed adenovirus-based DNA vaccines [171]. Therefore, our designed adenoviral DNA vaccine could be a potential candidate for Mtb control. Despite the promising results of the proposed vaccine, our study is based on computational subtractive proteomics and immunoinformatics approaches; we recommend laboratory validation to evaluate the efficacy of the proposed vaccine.
Experimental approaches can be time-consuming and laborious, often yielding minimal results. Consequently, bioinformatics approaches have become the preferred methods for scientists seeking to identify potential novel drug targets and vaccine candidates. This study utilized the subtractive proteomics approach to identify novel vaccine targets for 23 virulent Mtb strains. We ultimately selected a membrane protein, transmembrane transport protein MmpL4 NP_214964.1, as both a potential drug target and vaccine candidate. This protein, conserved in the other 22 virulent MDR and XDR strains of Mtb, demonstrated more than 99% sequence similarity. The pathogen-specific transmembrane transport protein MmpL4 is non-homologous to the host and essential for the pathogen's survival. MmpL4 proteins exhibited lower similarity with human gut microflora and were non-homologous to human anti-targets, thus being predicted as a novel therapeutic target. We further expanded our study to design a subunit vaccine from the MmpL4 protein using an immunoinformatics approach. The vaccine contained CTL, HTL, BCL epitopes, the 50s ribosomal protein L7/L12 of Mtb as a TLR-4 specific adjuvant, PADRE, and an invasin sequence to generate an appropriate immune response against tuberculosis infections. Therefore, this in silico study could save researchers both time and costs in finding effective solutions against infections caused by any of the 23 virulent MDR and XDR Mtb strains. This approach also reduces the need for extensive pre-clinical trials and repeated assay errors.
Notes
Acknowledgments
We would like to thank Md. Nahid Hasan, Ahsan Habib, and Ashikur Rahman Khan (student of genetic engineering and biotechnology, Shahjalal University of Science and Technology) for their help in the methodology section.
Supplementary Materials
Supplementary data can be found with this article online at http://www.genominfo.org.
Supplementary Table 1.
List of hypothetical proteins excluded in this study
Supplementary Table 2.
Antigenicity score and number of transmembrane helices
Supplementary Table 3.
List of 22 virulent strains of Mycobacterium tuberculosis
Supplementary Table 4.
Screening of cytotoxic T lymphocyte (CTL) epitopes
Supplementary Table 5.
Screening of helper T lymphocyte (HTL) epitopes
Supplementary Table 6.
Screening of B-cell lymphocyte (BCL) epitopes
Supplementary Table 7.
Quality validation of Three Dimensional (3D) structure of constructed vaccine and refined structures
Supplementary File 3.
List of accession numbers of 210 Anti-target proteins.
Fig. 1.
Overall protocol of subtractive proteomics and immunoinformatics approaches. Arrows indicate findings of subtractive proteomics are analyzed in immunoinformatics approach to design potential vaccine candidate.
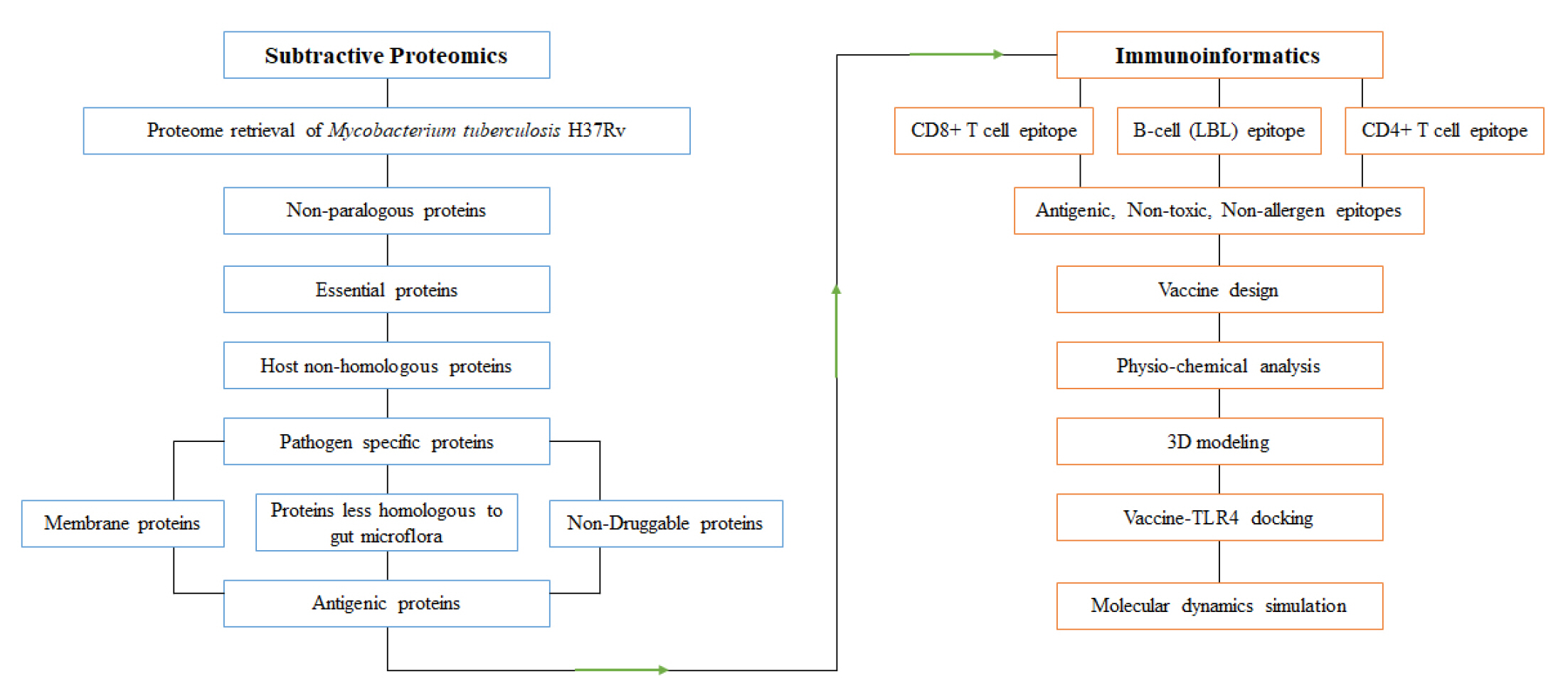
Fig. 2.
Protein-protein interaction analysis of ‘MmpL4’ protein. ‘MmpL4’ interacted with seven other proteins and among them two proteins ‘MmpS4’ and ‘MmpS5’ were involved in pathogenesis.
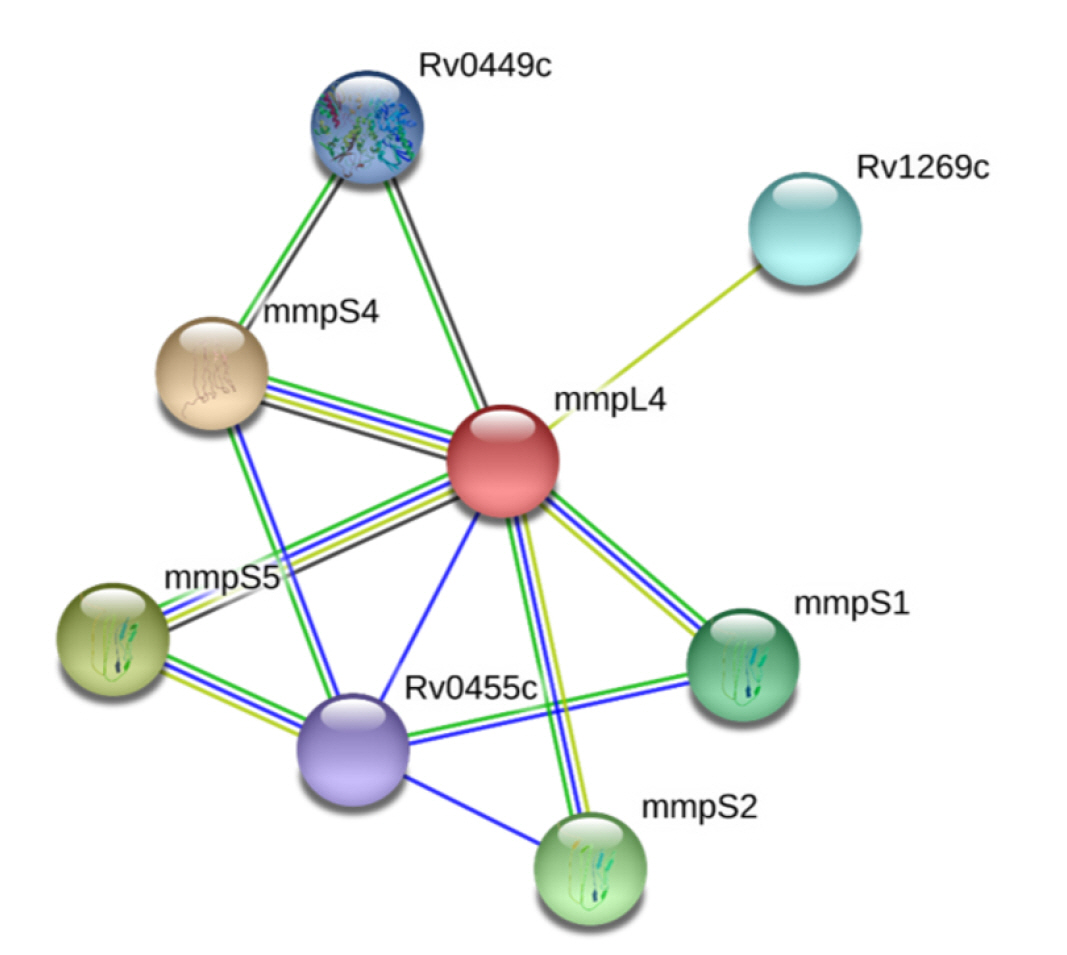
Fig. 3.
Worldwide population coverage analysis of selected cytotoxic T lymphocyte (CTL) and helper T lymphocyte (HTL) epitopes. CTL epitopes covered 98.84% MHC class I type HLA alleles worldwide whereas HTL covered 86.76% MHC class II type HLA alleles.

Fig. 4.
Vaccine construction, 3D structure design and refinement. (A) Selected epitopes were linearly joined with adjuvant (orange color), PADRE sequence (yellow color) and invasin peptide (ash color). Linker were used to separate each other. (B) A refined 3D structure of vaccine containing 443 amino acids. (C) PROCHECK tool revealed that 88.8% residues were present in most favored region and overall 99.5% residues were in allowed region.

Fig. 5.
Vaccine construction, 3D structure design and refinement. (A) Selected epitopes were linearly joined with adjuvant (orange color), PADRE sequence (yellow color) and invasin peptide (ash color). Linker were used to separate each other. (B) A refined 3D structure of vaccine containing 443 amino acids. (C) PROCHECK tool revealed that 88.8% residues were present in most favored region and overall 99.5% residues were in allowed region. RMSD, root mean square deviation; SASA, solvent accessible surface area; RMSF, root mean square fluctuation.
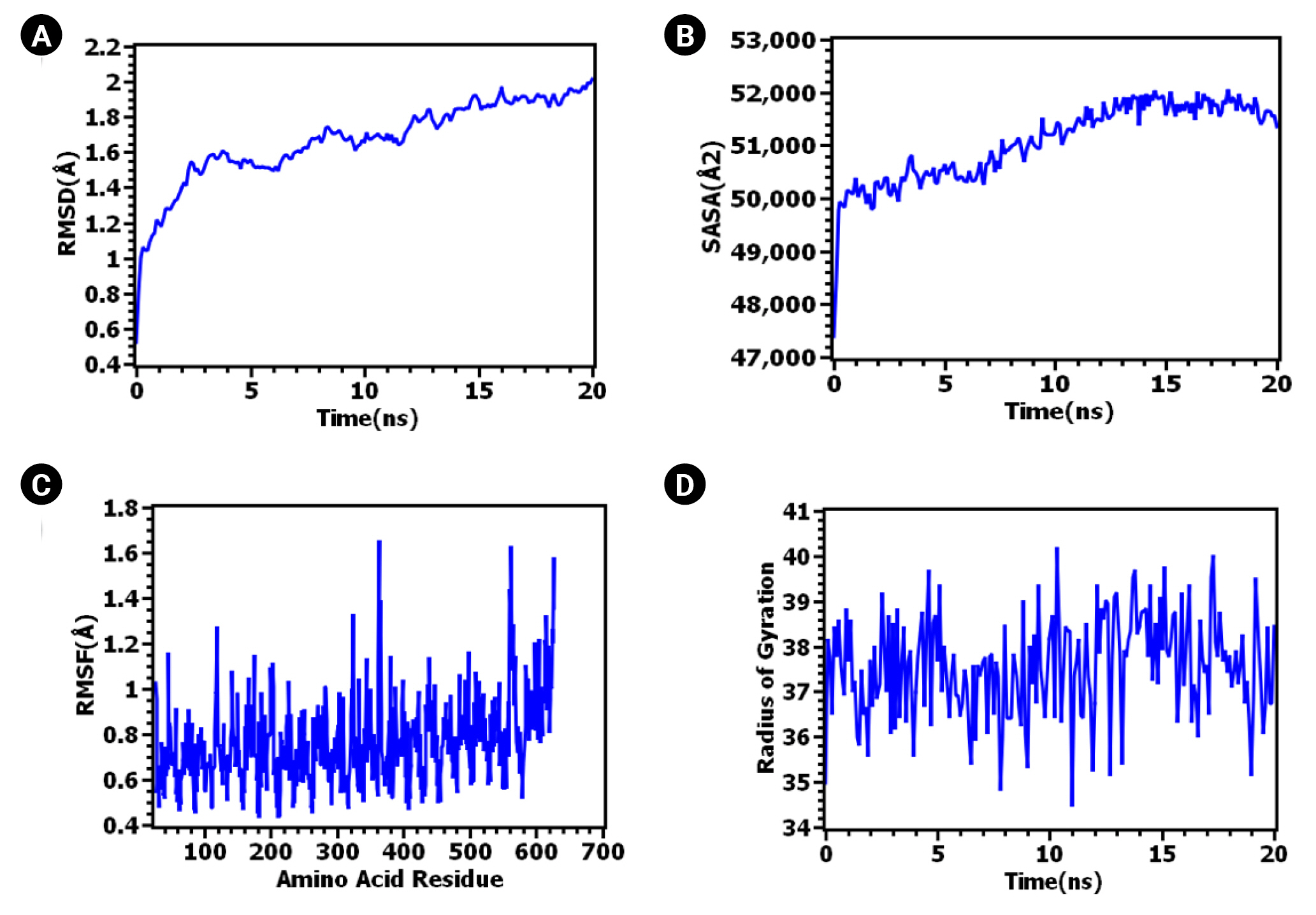
Fig. 6.
Protein-protein interactions of vaccine bound human Toll-like receptor-4 (TLR-4)/molecular dynamics (MD) complex. Vaccine molecule forms four hydrogen bonds with TLR-4 whereas two hydrogen bonds and one salt bridge interaction with myeloid differentiation factor-2. Rest of the contacts were non-bonded interactions.

Fig. 7.
Immune simulation analysis of designed vaccine over the course of three injections. (A) Response of immunoglobulin and immunocomplex to vaccine inoculations. (B) Rise in B-cell population. (C) Inclination of B-cell population. (D) Rise in plasma B cell. (E) Enhancement of helper T cell. (F) Rise in helper T cell. (G) Increase in regulatory T lymphocyte. (H) Augmentation in cytotoxic T lymphocyte. (I) Rise in active dendritic cell. (J) Increase in macrophages.
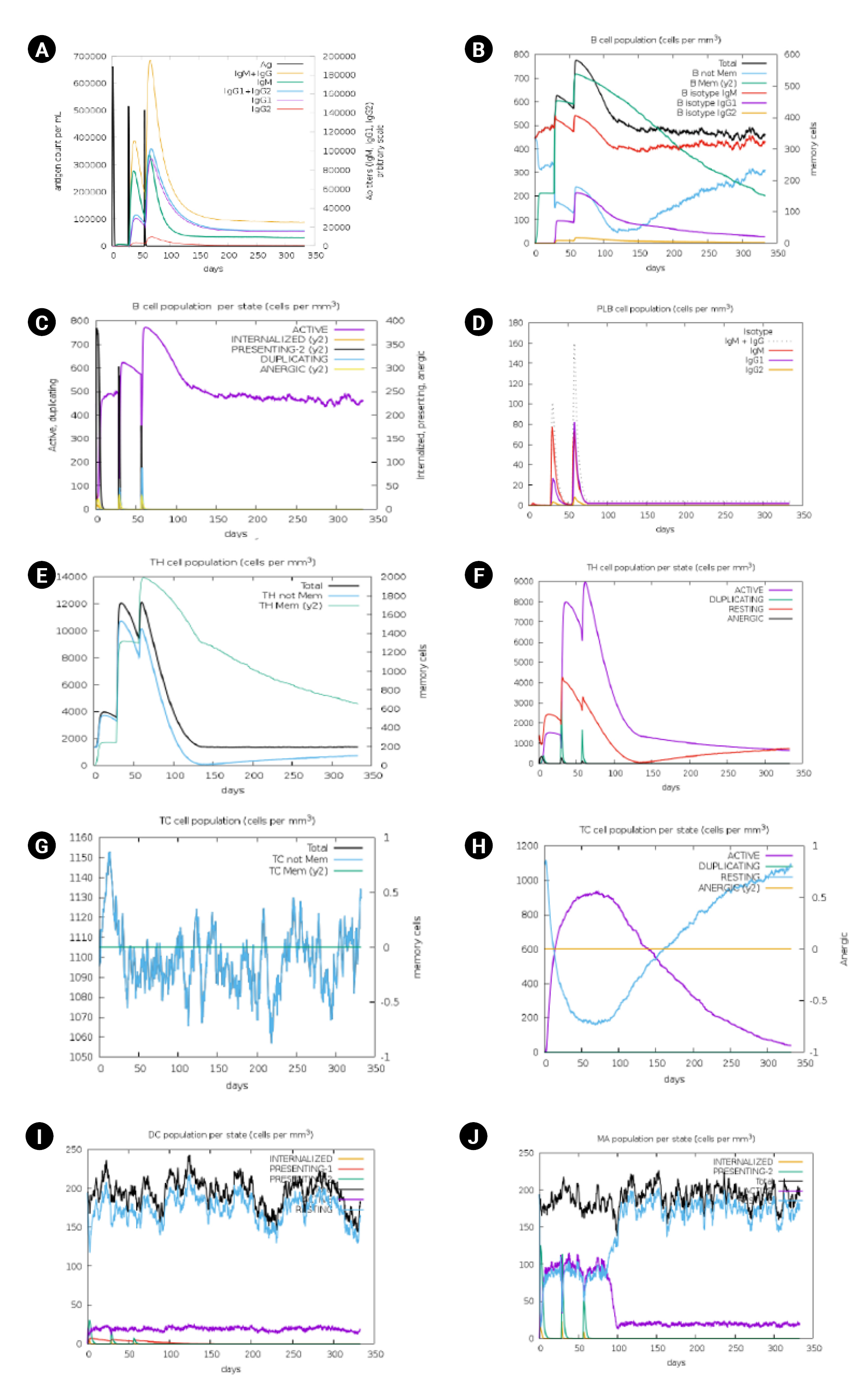
Table 1.
Subtractive proteomic analysis and its outcomes
Table 2.
Screening of 41 unique proteins based on localization, gut microflora non-homology, novelty, and molecular weight
Table 3.
Physiochemical properties, antigenicity, and allergenicity profiling of the constructed vaccine
Table 4.
Protein-protein interaction analysis after a 20-ns molecular dynamics simulation
References
1. World Health Organization. Global Tuberculosis Report 2020: Executive Summary. Geneva: World Health Organization, 2020.
2. World Health Organization. Global tuberculosis report 2019. Geneva: World Health Organization, 2019.
3. World Health Organization. Global tuberculosis report 2016. Geneva: World Health Organization, 2016.
4. Tuberculosis: multidrug-resistant tuberculosis (MDR-TB). Geneva: World Health Organization, 2018. [2021 Aug 8]. Available from: https://www.who.int/news-room/q-a-detail/tuberculosis-multidrug-resistant-tuberculosis-(mdr-tb).
5. Nguyen QH, Contamin L, Nguyen TV, Banuls AL. Insights into the processes that drive the evolution of drug resistance in Mycobacterium tuberculosis. Evol Appl 2018;11:1498–1511.




6. World Health Organization. WHO treatment guidelines for drug-resistant tuberculosis. Geneva: World Health Organization, 2016.
7. Hema K, Priyadarshini VI, Pradhan D, Munikumar M, Sandeep S, Pradeep N, et al. Identification of putative drug targets and vaccine candidates for pathogens causing atherosclerosis. Biochem Anal Biochem 2015;4:175.

8. Almeida Da Silva PE, Palomino JC. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother 2011;66:1417–1430.


9. Sharma R, Rajput VS, Jamal S, Grover A, Grover S. An immunoinformatics approach to design a multi-epitope vaccine against Mycobacterium tuberculosis exploiting secreted exosome proteins. Sci Rep 2021;11:13836.




10. Cohen KA, Manson AL, Desjardins CA, Abeel T, Earl AM. Deciphering drug resistance in Mycobacterium tuberculosis using whole-genome sequencing: progress, promise, and challenges. Genome Med 2019;11:45.




11. Dookie N, Rambaran S, Padayatchi N, Mahomed S, Naidoo K. Evolution of drug resistance in Mycobacterium tuberculosis: a review on the molecular determinants of resistance and implications for personalized care. J Antimicrob Chemother 2018;73:1138–1151.



12. Buchy P, Ascioglu S, Buisson Y, Datta S, Nissen M, Tambyah PA, et al. Impact of vaccines on antimicrobial resistance. Int J Infect Dis 2020;90:188–196.


13. Bloom DE, Black S, Salisbury D, Rappuoli R. Antimicrobial resistance and the role of vaccines. Proc Natl Acad Sci U S A 2018;115:12868–12871.



14. Xing Z, Charters TJ. Heterologous boost vaccines for bacillus Calmette-Guerin prime immunization against tuberculosis. Expert Rev Vaccines 2007;6:539–546.

15. Parida SK, Kaufmann SH. Novel tuberculosis vaccines on the horizon. Curr Opin Immunol 2010;22:374–384.


16. Horvath CN, Xing Z. Immunization strategies against pulmonary tuberculosis: considerations of T cell geography. Adv Exp Med Biol 2013;783:267–278.


17. Hussey G, Hawkridge T, Hanekom W. Childhood tuberculosis: old and new vaccines. Paediatr Respir Rev 2007;8:148–154.


18. Zhang W, Zhang Y, Zheng H, Pan Y, Liu H, Du P, et al. Genome sequencing and analysis of BCG vaccine strains. PLoS One 2013;8:e71243.



19. Singh SP, Mishra BN. Major histocompatibility complex linked databases and prediction tools for designing vaccines. Hum Immunol 2016;77:295–306.


20. Hanekom M, Gey van Pittius NC, McEvoy C, Victor TC, Van Helden PD, Warren RM. Mycobacterium tuberculosis Beijing genotype: a template for success. Tuberculosis (Edinb) 2011;91:510–523.


21. Mendez-Samperio P. Novel vaccination strategies and approaches against human tuberculosis. Scand J Immunol 2019;90:e12774.

22. Marciano BE, Huang CY, Joshi G, Rezaei N, Carvalho BC, Allwood Z, et al. BCG vaccination in patients with severe combined immunodeficiency: complications, risks, and vaccination policies. J Allergy Clin Immunol 2014;133:1134–1141.



23. Barkai G, Somech R, Stauber T, Barziali A, Greenberger S. Bacille Calmette-Guerin (BCG) complications in children with severe combined immunodeficiency (SCID). Infect Dis (Lond) 2019;51:585–592.


24. Nunes-Santos CJ, Rosenzweig SD. Bacille Calmette-Guerin complications in newly described primary immunodeficiency diseases: 2010-2017. Front Immunol 2018;9:1423.



25. Sable SB, Posey JE, Scriba TJ. Tuberculosis vaccine development: progress in clinical evaluation. Clin Microbiol Rev 2019;33:e00100–00119.




26. Soundarya JS, Ranganathan UD, Tripathy SP. Current trends in tuberculosis vaccine. Med J Armed Forces India 2019;75:18–24.



27. Martin C, Aguilo N, Marinova D, Gonzalo-Asensio JJ. Update on TB vaccine pipeline. Appl Sci 2020;10:2632.

28. Solanki V, Tiwari V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci Rep 2018;8:9044.




29. de Sarom A, Kumar Jaiswal A, Tiwari S, de Castro Oliveira L, Barh D, Azevedo V, et al. Putative vaccine candidates and drug targets identified by reverse vaccinology and subtractive genomics approaches to control Haemophilus ducreyi, the causative agent of chancroid. J R Soc Interface 2018;15:20180032.




30. Mahmud A, Khan MT, Iqbal A. Identification of novel drug targets for humans and potential vaccine targets for cattle by subtractive genomic analysis of Brucella abortus strain 2308. Microb Pathog 2019;137:103731.


31. Khan MT, Mahmud A, Iqbal A, Hoque SF, Hasan M. Subtractive genomics approach towards the identification of novel therapeutic targets against human Bartonella bacilliformis. Inf Med Inlocked 2020;20:100385.

32. Khan MT, Islam MJ, Parihar A, Islam R, Jerin TJ, Dhote R, et al. Immunoinformatics and molecular modeling approach to design universal multi-epitope vaccine for SARS-CoV-2. Inform Med Unlocked 2021;24:100578.



33. Khan MT, Islam R, Jerin TJ, Mahmud A, Khatun S, Kobir A, et al. Immunoinformatics and molecular dynamics approaches: next generation vaccine design against West Nile virus. PLoS One 2021;16:e0253393.



34. Parvizpour S, Pourseif MM, Razmara J, Rafi MA, Omidi Y. Epitope-based vaccine design: a comprehensive overview of bioinformatics approaches. Drug Discov Today 2020;25:1034–1042.


35. Kazi A, Chuah C, Majeed ABA, Leow CH, Lim BH, Leow CY. Current progress of immunoinformatics approach harnessed for cellular- and antibody-dependent vaccine design. Pathog Glob Health 2018;112:123–131.



36. Calis JJ, Maybeno M, Greenbaum JA, Weiskopf D, De Silva AD, Sette A, et al. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput Biol 2013;9:e1003266.



37. Zhang L. Multi-epitope vaccines: a promising strategy against tumors and viral infections. Cell Mol Immunol 2018;15:182–184.



38. Zhu S, Feng Y, Rao P, Xue X, Chen S, Li W, et al. Hepatitis B virus surface antigen as delivery vector can enhance Chlamydia trachomatis MOMP multi-epitope immune response in mice. Appl Microbiol Biotechnol 2014;98:4107–4117.



39. Jiang P, Cai Y, Chen J, Ye X, Mao S, Zhu S, et al. Evaluation of tandem Chlamydia trachomatis MOMP multi-epitopes vaccine in BALB/c mice model. Vaccine 2017;35:3096–3103.


40. Pumchan A, Krobthong S, Roytrakul S, Sawatdichaikul O, Kondo H, Hirono I, et al. Novel chimeric multiepitope vaccine for streptococcosis disease in Nile Tilapia (Oreochromis niloticus Linn.). Sci Rep 2020;10:603.




41. Caro-Gomez E, Gazi M, Goez Y, Valbuena G. Discovery of novel cross-protective Rickettsia prowazekii T-cell antigens using a combined reverse vaccinology and in vivo screening approach. Vaccine 2014;32:4968–4976.



42. Lin X, Chen S, Xue X, Lu L, Zhu S, Li W, et al. Chimerically fused antigen rich of overlapped epitopes from latent membrane protein 2 (LMP2) of Epstein-Barr virus as a potential vaccine and diagnostic agent. Cell Mol Immunol 2016;13:492–501.



43. Yusufu M, Shalitanati A, Yu H, Moming A, Li Y, Deng F, et al. Immune responses in mice induced by multi-epitope DNA vaccine and protein vaccine of Crimean-Congo hemorrhagic fever virus. Preprint at: https://doi.org/10.1101/719724 (2019).

44. Foroutan M, Ghaffarifar F, Sharifi Z, Dalimi A. Vaccination with a novel multi-epitope ROP8 DNA vaccine against acute Toxoplasma gondii infection induces strong B and T cell responses in mice. Comp Immunol Microbiol Infect Dis 2020;69:101413.


45. Jasenosky LD, Scriba TJ, Hanekom WA, Goldfeld AE. T cells and adaptive immunity to Mycobacterium tuberculosis in humans. Immunol Rev 2015;264:74–87.


46. Lewinsohn DA, Lewinsohn DM, Scriba TJ. Polyfunctional CD4(+) T cells as targets for tuberculosis vaccination. Front Immunol 2017;8:1262.



47. Maglione PJ, Xu J, Chan J. B cells moderate inflammatory progression and enhance bacterial containment upon pulmonary challenge with Mycobacterium tuberculosis. J Immunol 2007;178:7222–7234.



48. Kozakiewicz L, Phuah J, Flynn J, Chan J. The role of B cells and humoral immunity in Mycobacterium tuberculosis infection. Adv Exp Med Biol 2013;783:225–250.



49. Melly G, Purdy GE. MmpL proteins in physiology and pathogenesis of M. tuberculosis. Microorganisms 2019;7:70.



50. Szekely R, Cole ST. Mechanistic insight into mycobacterial MmpL protein function. Mol Microbiol 2016;99:831–834.



51. Wells RM, Jones CM, Xi Z, Speer A, Danilchanka O, Doornbos KS, et al. Discovery of a siderophore export system essential for virulence of Mycobacterium tuberculosis. PLoS Pathog 2013;9:e1003120.



52. Jones CM, Niederweis M. Mycobacterium tuberculosis can utilize heme as an iron source. J Bacteriol 2011;193:1767–1770.




53. Domenech P, Reed MB, Barry CE 3rd. Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect Immun 2005;73:3492–3501.




54. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998;393:537–544.



55. Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 2010;26:680–682.




56. Uddin R, Azam SS, Wadood A, Khan W, Farooq U, Khan A. Computational identification of potential drug targets against Mycobacterium leprae. Med Chem Res 2016;25:473–481.


57. Gupta SK, Sarita S, Gupta MK, Pant KK, Seth PK. Definition of potential targets in Mycoplasma pneumoniae through subtractive genome analysis. J Antivir Antiretrovir 2010;2:38–41.
58. Luo H, Lin Y, Gao F, Zhang CT, Zhang R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res 2014;42:D574–D580.

59. Butt AM, Tahir S, Nasrullah I, Idrees M, Lu J, Tong Y. Mycoplasma genitalium: a comparative genomics study of metabolic pathways for the identification of drug and vaccine targets. Infect Genet Evol 2012;12:53–62.


60. Mondal SI, Ferdous S, Jewel NA, Akter A, Mahmud Z, Islam MM, et al. Identification of potential drug targets by subtractive genome analysis of Escherichia coli O157:H7: an in silico approach. Adv Appl Bioinform Chem 2015;8:49–63.



61. Murali S, Jahageerdar S, Kumar S, Krishna G. Computational identification and screening of natural compounds as drug targets against the fish pathogen, Pseudomonas fluorescens. Int J Curr Microbiol App Sci 2017;6:3521–3535.

62. Rahman A, Noore S, Hasan A, Ullah R, Rahman H, Hossain A, et al. Identification of potential drug targets by subtractive genome analysis of Bacillus anthracis A0248: an in silico approach. Comput Biol Chem 2014;52:66–72.


63. Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, et al. Ensembl 2018. Nucleic Acids Res 2018;46:D754–D761.


64. Hossain M, Chowdhury DU, Farhana J, Akbar MT, Chakraborty A, Islam S, et al. Identification of potential targets in Staphylococcus aureus N315 using computer aided protein data analysis. Bioinformation 2013;9:187–192.



65. Trivedi G, Georrge JJ. Identification of novel drug targets and its inhibitor from essential genes of human pathogenic gam positive bacteria. In: 9th National Level Science Symposium, 2016 Feb 14; Rajkot, India: pp 314–319.
66. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30.



67. Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 2007;35:W182–W185.



68. Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins 2006;64:643–651.


69. Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010;26:1608–1615.




70. Bhasin M, Garg A, Raghava GP. PSLpred: prediction of subcellular localization of bacterial proteins. Bioinformatics 2005;21:2522–2524.


71. Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46:D1074–D1082.


72. Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, et al. Protein identification and analysis tools on the ExPASy server. In: The Proteomics Protocols Handbook (Walker JM, ed.). Dordrecht: Springer, 2005. pp. 571–607.
73. Shanmugham B, Pan A. Identification and characterization of potential therapeutic candidates in emerging human pathogen Mycobacterium abscessus: a novel hierarchical in silico approach. PLoS One 2013;8:e59126.



74. Raman K, Yeturu K, Chandra N. targetTB: a target identification pipeline for Mycobacterium tuberculosis through an interactome, reactome and genome-scale structural analysis. BMC Syst Biol 2008;2:109.




75. Doytchinova IA, Flower DR. Identifying candidate subunit vaccines using an alignment-independent method based on principal amino acid properties. Vaccine 2007;25:856–866.


76. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001;305:567–580.

77. Nalamolu RM, Pasala C, Katari SK, Amineni U. Discovery of common putative drug targets and vaccine candidates for Mycobacterium tuberculosis sp. J Drug Deliv Ther 2019;9:67–71.


78. Sridhar S, Dash P, Guruprasad K. Comparative analyses of the proteins from Mycobacterium tuberculosis and human genomes: identification of potential tuberculosis drug targets. Gene 2016;579:69–74.


79. Arifuzzaman M, Maeda M, Itoh A, Nishikata K, Takita C, Saito R, et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res 2006;16:686–691.



80. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–D613.


81. Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot. Methods Mol Biol 2007;406:89–112.


82. Usmani SS, Kumar R, Bhalla S, Kumar V, Raghava GPS. In silico tools and databases for designing peptide-based vaccine and drugs. Adv Protein Chem Struct Biol 2018;112:221–263.


83. Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics 2007;8:424.




84. Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, et al. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics 2010;11:568.




85. Saha S, Raghava GP. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins 2006;65:40–48.


86. Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res 2017;45:W24–W29.



87. Dimitrov I, Flower DR, Doytchinova I. AllerTOP: a server for in silico prediction of allergens. BMC Bioinformatics 2013;14 Suppl 6:S4.



88. Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, Open Source Drug Discovery Consortium, et al. In silico approach for predicting toxicity of peptides and proteins. PLoS One 2013;8:e73957.



89. Dhanda SK, Vir P, Raghava GP. Designing of interferon-gamma inducing MHC class-II binders. Biol Direct 2013;8:30.




90. Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics 2006;7:153.




91. Pandey RK, Ali M, Ojha R, Bhatt TK, Prajapati VK. Development of multi-epitope driven subunit vaccine in secretory and membrane protein of Plasmodium falciparum to convey protection against malaria infection. Vaccine 2018;36:4555–4565.


92. Lee SJ, Shin SJ, Lee MH, Lee MG, Kang TH, Park WS, et al. A potential protein adjuvant derived from Mycobacterium tuberculosis Rv0652 enhances dendritic cells-based tumor immunotherapy. PLoS One 2014;9:e104351.



93. Khan MT, Mahmud A, Hasan M, Azim KF, Begum MK, Akter A, et al. Proteome exploration of Legionella pneumophila for identifying novel therapuetics: a hierarchical subtractive genomics and reverse vaccinology approach. Preprint at: https://doi.org/10.1101/2020.02.03.922864 (2020).

94. Rahman MS, Hoque MN, Islam MR, Akter S, Rubayet Ul Alam A, Siddique MA, et al. Epitope-based chimeric peptide vaccine design against S, M and E proteins of SARS-CoV-2 etiologic agent of global pandemic COVID-19: an in silico approach. PeerJ 2020;8:e9572.




95. Saha S, Raghava GP. AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res 2006;34:W202–W209.



96. Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 2004;32:W526–W531.



97. Heo L, Park H, Seok C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res 2013;41:W384–W388.



98. Laskowski RA, MacArthur MW, Moss DS, Thornton JM. ProCheck: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 1993;26:283–291.

99. Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature 1992;356:83–85.



100. Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 2007;35:W407–W410.



101. Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res 2004;32:W96–W99.



102. Krieger E, Darden T, Nabuurs SB, Finkelstein A, Vriend G. Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins 2004;57:678–683.


103. Krieger E, Nielsen JE, Spronk CA, Vriend G. Fast empirical pKa prediction by Ewald summation. J Mol Graph Model 2006;25:481–486.


104. Rapin N, Lund O, Bernaschi M, Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS One 2010;5:e9862.



105. Samad A, Meghla NS, Nain Z, Karpinski TM, Rahman MS. Immune epitopes identification and designing of a multi-epitope vaccine against bovine leukemia virus: a molecular dynamics and immune simulation approaches. Cancer Immunol Immunother 2022;71:2535–2548.




106. Sarkar M, Maganti L, Ghoshal N, Dutta C. In silico quest for putative drug targets in Helicobacter pylori HPAG1: molecular modeling of candidate enzymes from lipopolysaccharide biosynthesis pathway. J Mol Model 2012;18:1855–1866.



107. Hadizadeh M, Tabatabaiepour SN, Tabatabaiepour SZ, Hosseini Nave H, Mohammadi M, Sohrabi SM. Genome-wide identification of potential drug target in Enterobacteriaceae family: a homology-based method. Microb Drug Resist 2018;24:8–17.


108. Aguero F, Al-Lazikani B, Aslett M, Berriman M, Buckner FS, Campbell RK, et al. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat Rev Drug Discov 2008;7:900–907.




109. Butt AM, Nasrullah I, Tahir S, Tong Y. Comparative genomics analysis of Mycobacterium ulcerans for the identification of putative essential genes and therapeutic candidates. PLoS One 2012;7:e43080.



110. Damte D, Suh JW, Lee SJ, Yohannes SB, Hossain MA, Park SC. Putative drug and vaccine target protein identification using comparative genomic analysis of KEGG annotated metabolic pathways of Mycoplasma hyopneumoniae. Genomics 2013;102:47–56.


111. Kovatcheva-Datchary P, Zoetendal EG, Venema K, de Vos WM, Smidt H. Tools for the tract: understanding the functionality of the gastrointestinal tract. Therap Adv Gastroenterol 2009;2:9–22.


112. Hooper LV, Bry L, Falk PG, Gordon JI. Host-microbial symbiosis in the mammalian intestine: exploring an internal ecosystem. Bioessays 1998;20:336–343.


115. Recanatini M, Bottegoni G, Cavalli A. In silico antitarget screening. Drug Discov Today Technol 2004;1:209–215.


116. Fung M, Thornton A, Mybeck K, Wu JH, Hornbuckle K, Muniz E. Evaluation of the characteristics of safety withdrawal of prescription drugs from worldwide pharmaceutical markets-1960 to 1999. Drug Inf J 2001;35:293–317.


117. Fenner H. Evaluation of the efficacy and safety of NSAIDs. A new methodological approach. Scand J Rheumatol Suppl 1989;80:32–39.


118. Shaw PJ, Ganey PE, Roth RA. Idiosyncratic drug-induced liver injury and the role of inflammatory stress with an emphasis on an animal model of trovafloxacin hepatotoxicity. Toxicol Sci 2010;118:7–18.



120. Birhanu BT, Lee SJ, Park NH, Song JB, Park SC. In silico analysis of putative drug and vaccine targets of the metabolic pathways of Actinobacillus pleuropneumoniae using a subtractive/comparative genomics approach. J Vet Sci 2018;19:188–199.




121. Uddin R, Siddiqui QN, Azam SS, Saima B, Wadood A. Identification and characterization of potential druggable targets among hypothetical proteins of extensively drug resistant Mycobacterium tuberculosis (XDR KZN 605) through subtractive genomics approach. Eur J Pharm Sci 2018;114:13–23.


122. Uddin R, Zahra NU, Azam SS. Identification of glucosyl-3-phosphoglycerate phosphatase as a novel drug target against resistant strain of Mycobacterium tuberculosis (XDR1219) by using comparative metabolic pathway approach. Comput Biol Chem 2019;79:91–102.


123. Dar HA, Zaheer T, Ullah N, Bakhtiar SM, Zhang T, Yasir M, et al. Pangenome analysis of Mycobacterium tuberculosis reveals core-drug targets and screening of promising lead compounds for drug discovery. Antibiotics (Basel) 2020;9:819.



124. Gupta D, Banerjee S, Pailan S, Saha P. In silico identification and characterization of a hypothetical protein of Mycobacterium tuberculosis EAI5 as a potential virulent factor. Bioinformation 2016;12:182–191.



125. Nain Z, Karim MM, Sen MK, Adhikari UK. Structural basis and designing of peptide vaccine using PE-PGRS family protein of Mycobacterium ulcerans: an integrated vaccinomics approach. Mol Immunol 2020;120:146–163.


126. Li N, Liu P, Wang L, Liu J, Yuan X, Meng W, et al. Effect of Ipr1 on expression levels of immune genes related to macrophage anti-infection of Mycobacterium tuberculosis. Int J Clin Exp Med 2015;8:3411–3419.


127. Choi HG, Kim WS, Back YW, Kim H, Kwon KW, Kim JS, et al. Mycobacterium tuberculosis RpfE promotes simultaneous Th1- and Th17-type T-cell immunity via TLR4-dependent maturation of dendritic cells. Eur J Immunol 2015;45:1957–1971.


128. Pivarcsi A, Bodai L, Rethi B, Kenderessy-Szabo A, Koreck A, Szell M, et al. Expression and function of Toll-like receptors 2 and 4 in human keratinocytes. Int Immunol 2003;15:721–730.


129. Abel B, Thieblemont N, Quesniaux VJ, Brown N, Mpagi J, Miyake K, et al. Toll-like receptor 4 expression is required to control chronic Mycobacterium tuberculosis infection in mice. J Immunol 2002;169:3155–3162.



130. Zare-Bidaki M, Hakimi H, Abdollahi SH, Zainodini N, Arababadi MK, Kennedy D. TLR4 in toxoplasmosis; friends or foe? Microb Pathog 2014;69-70:28–32.


131. Afsharimoghaddam A, Soleimani M, Lashay A, Dehghani M, Sepehri Z. Controversial roles played by toll like receptor 4 in urinary bladder cancer: a systematic review. Life Sci 2016;158:31–36.


132. Sepehri Z, Kiani Z, Kohan F, Ghavami S. Toll-like receptor 4 as an immune receptor against Mycobacterium tuberculosis: a systematic review. Lab Med 2019;50:117–129.


133. Islam MJ, Parves MR, Mahmud S, Tithi FA, Reza MA. Assessment of structurally and functionally high-risk nsSNPs impacts on human bone morphogenetic protein receptor type IA (BMPR1A) by computational approach. Comput Biol Chem 2019;80:31–45.


134. Mahmud S, Parves MR, Riza YM, Sujon KM, Ray S, Tithi FA, et al. Exploring the potent inhibitors and binding modes of phospholipase A2 through in silico investigation. J Biomol Struct Dyn 2020;38:4221–4231.


135. Jeffrey GA. An introduction to hydrogen bonding. New York: Oxford university Press, 1997.
136. Hendsch ZS, Tidor B. Do salt bridges stabilize proteins? A continuum electrostatic analysis. Protein Sci 1994;3:211–226.




137. Bosshard HR, Marti DN, Jelesarov I. Protein stabilization by salt bridges: concepts, experimental approaches and clarification of some misunderstandings. J Mol Recognit 2004;17:1–16.


138. Makhatadze GI, Loladze VV, Ermolenko DN, Chen X, Thomas ST. Contribution of surface salt bridges to protein stability: guidelines for protein engineering. J Mol Biol 2003;327:1135–1148.


139. Kennedy DA, Read AF. Why the evolution of vaccine resistance is less of a concern than the evolution of drug resistance. Proc Natl Acad Sci U S A 2018;115:12878–12886.



140. Relman DA, Lipsitch M. Microbiome as a tool and a target in the effort to address antimicrobial resistance. Proc Natl Acad Sci U S A 2018;115:12902–12910.



141. Plotkin S, Robinson JM, Cunningham G, Iqbal R, Larsen S. The complexity and cost of vaccine manufacturing: an overview. Vaccine 2017;35:4064–4071.



142. Nguyen Thi LT, Sarmiento ME, Calero R, Camacho F, Reyes F, Hossain MM, et al. Immunoinformatics study on highly expressed Mycobacterium tuberculosis genes during infection. Tuberculosis (Edinb) 2014;94:475–481.


143. Rahmat Ullah S, Majid M, Rashid MI, Mehmood K, Andleeb S. Immunoinformatics driven prediction of multiepitopic vaccine against Klebsiella pneumoniae and Mycobacterium tuberculosis coinfection and its validation via in silico expression. Int J Pept Res Ther 2021;27:987–999.



144. Prezzemolo T, Guggino G, La Manna MP, Di Liberto D, Dieli F, Caccamo N. Functional signatures of human CD4 and CD8 T cell responses to Mycobacterium tuberculosis. Front Immunol 2014;5:180.



145. Lin PL, Flynn JL. CD8 T cells and Mycobacterium tuberculosis infection. Semin Immunopathol 2015;37:239–249.




146. Stenger S, Hanson DA, Teitelbaum R, Dewan P, Niazi KR, Froelich CJ, et al. An antimicrobial activity of cytolytic T cells mediated by granulysin. Science 1998;282:121–125.


147. Serbina NV, Liu CC, Scanga CA, Flynn JL. CD8+ CTL from lungs of Mycobacterium tuberculosis-infected mice express perforin in vivo and lyse infected macrophages. J Immunol 2000;165:353–363.



148. Cho S, Mehra V, Thoma-Uszynski S, Stenger S, Serbina N, Mazzaccaro RJ, et al. Antimicrobial activity of MHC class I-restricted CD8+ T cells in human tuberculosis. Proc Natl Acad Sci U S A 2000;97:12210–12215.



150. North RJ. Importance of thymus-derived lymphocytes in cell-mediated immunity to infection. Cell Immunol 1973;7:166–176.


151. Lefford MJ. Transfer of adoptive immunity to tuberculosis in mice. Infect Immun 1975;11:1174–1181.




152. Khan TA, Mazhar H, Saleha S, Tipu HN, Muhammad N, Abbas MN. Interferon-gamma improves macrophages function against M. tuberculosis in multidrug-resistant tuberculosis patients. Chemother Res Pract 2016;2016:7295390.




153. Geginat J, Sallusto F, Lanzavecchia A. Cytokine-driven proliferation and differentiation of human naive, central memory and effector memory CD4+ T cells. Pathol Biol (Paris) 2003;51:64–66.



154. Walzl G, Ronacher K, Hanekom W, Scriba TJ, Zumla A. Immunological biomarkers of tuberculosis. Nat Rev Immunol 2011;11:343–354.



155. Rozot V, Vigano S, Mazza-Stalder J, Idrizi E, Day CL, Perreau M, et al. Mycobacterium tuberculosis-specific CD8+ T cells are functionally and phenotypically different between latent infection and active disease. Eur J Immunol 2013;43:1568–1577.



156. Brighenti S, Andersson J. Local immune responses in human tuberculosis: learning from the site of infection. J Infect Dis 2012;205 Suppl 2:S316–324.


157. Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med 1993;178:2249–2254.




158. Nandi B, Behar SM. Regulation of neutrophils by interferon-gamma limits lung inflammation during tuberculosis infection. J Exp Med 2011;208:2251–2262.


159. Chan J, Tanaka K, Carroll D, Flynn J, Bloom BR. Effects of nitric oxide synthase inhibitors on murine infection with Mycobacterium tuberculosis. Infect Immun 1995;63:736–740.




160. Dalton DK, Pitts-Meek S, Keshav S, Figari IS, Bradley A, Stewart TA. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science 1993;259:1739–1742.


161. Rao M, Valentini D, Poiret T, Dodoo E, Parida S, Zumla A, et al. B in TB: B cells as mediators of clinically relevant immune responses in tuberculosis. Clin Infect Dis 2015;61Suppl 3:S225–S234.


162. Kringelum JV, Nielsen M, Padkjaer SB, Lund O. Structural analysis of B-cell epitopes in antibody:protein complexes. Mol Immunol 2013;53:24–34.


163. Porada CD, Almeida-Porada G. Mesenchymal stem cells as therapeutics and vehicles for gene and drug delivery. Adv Drug Deliv Rev 2010;62:1156–1166.



164. Li X, Yang X, Jiang Y, Liu J. A novel HBV DNA vaccine based on T cell epitopes and its potential therapeutic effect in HBV transgenic mice. Int Immunol 2005;17:1293–1302.


165. Li H, Ning P, Lin Z, Liang W, Kang K, He L, et al. Co-expression of the C-terminal domain of Yersinia enterocolitica invasin enhances the efficacy of classical swine-fever-vectored vaccine based on human adenovirus. J Biosci 2015;40:79–90.



166. Xiang K, Ying G, Yan Z, Shanshan Y, Lei Z, Hongjun L, et al. Progress on adenovirus-vectored universal influenza vaccines. Hum Vaccin Immunother 2015;11:1209–1222.



167. Kallel H, Kamen AA. Large-scale adenovirus and poxvirus-vectored vaccine manufacturing to enable clinical trials. Biotechnol J 2015;10:741–747.


168. Farnos O, Gelaye E, Trabelsi K, Bernier A, Subramani K, Kallel H, et al. Establishing a Robust manufacturing platform for recombinant veterinary vaccines: an adenovirus-vector vaccine to control newcastle disease virus infections of poultry in sub-Saharan Africa. Vaccines (Basel) 2020;8:338.



169. Cai X, Bai H, Zhang X. Vaccines and advanced vaccines: a landscape for advanced vaccine technology against infectious disease, COVID-19 and tumor. Preprint at: https://doi.org/10.31219/osf.io/ypgx4 (2020).

170. Milligan ID, Gibani MM, Sewell R, Clutterbuck EA, Campbell D, Plested E, et al. Safety and immunogenicity of novel adenovirus type 26- and modified vaccinia Ankara-vectored Ebola vaccines: a randomized clinical trial. JAMA 2016;315:1610–1623.


171. Zimmer C, Corum J, Wee SL, Kristoggersen M. Coronavirus Vaccine Tracker. New York: The New York Times, 2022. [cited 2023 Mar 28]. Available from: https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html.
- TOOLS
-
METRICS

-
- 2 Crossref
- 0 Scopus
- 3,648 View
- 108 Download








