 |
 |
- Search
| Genomics Inform > Volume 21(3); 2023 > Article |
|
Abstract
The Bacillus cereus group, also known as B. cereus sensu lato (B. cereus s.l.), is composed of various Bacillus species, some of which can cause diarrheal or emetic food poisoning. Several emerging highly heat-resistant Bacillus species have been identified, these include B. thermoamylovorans, B. sporothermodurans, and B. cytotoxicus NVH 391-98. Herein, we performed whole genome analysis of two thermotolerant Bacillus sp. isolates, Bacillus sp. B48 and Bacillus sp. B140, from an omelet with acacia leaves and fried rice, respectively. Phylogenomic analysis suggested that Bacillus sp. B48 and Bacillus sp. B140 are closely related to B. cereus and B. thuringiensis, respectively. Whole genome alignment of Bacillus sp. B48, Bacillus sp. B140, mesophilic strain B. cereus ATCC14579, and thermophilic strain B. cytotoxicus NVH 391-98 using the Mauve program revealed the presence of numerous homologous regions including genes responsible for heat shock in the dnaK gene cluster. However, the presence of a DUF4253 domain-containing protein was observed only in the genome of B. cereus ATCC14579 while the intracellular protease PfpI family was present only in the chromosome of B. cytotoxicus NVH 391-98. In addition, prophage Clp protease-like proteins were found in the genomes of both Bacillus sp. B48 and Bacillus sp. B140 but not in the genome of B. cereus ATCC14579. The genomic profiles of Bacillus sp. isolates were identified by using whole genome analysis especially those relating to heat-responsive gene clusters. The findings presented in this study lay the foundations for subsequent studies to reveal further insights into the molecular mechanisms of Bacillus species in terms of heat resistance mechanisms.
Bacillus cereus is a spore-forming gram-positive rod that is a common contaminant of food and dairy products. B. cereus is one of the major foodborne pathogens that is responsible for causing diarrheal or emetic food poisoning. B. cereus strains can be divided into distinct groups based on their growth and survival characteristics (i.e., psychrophilic, mesophilic, and thermophilic). Mesophilic strains grow well at 37ºC and can survive at temperatures below 10ºC [1]. In contrast, psychrophilic strains grow effectively at temperatures below 10ºC, but grow poorly at 37ºC [1]. Psychrophilic strains of B. cereus are commonly present in chilled or fresh foods [2-4]. However, some B. cereus strains can grow between 20–50ºC and this is representative of a cluster of thermophilic strains [5]. B. cereus can resist heat, dryness, and disinfectants since it is able to produce heat-resistant spores [6]. These spores may subsequently germinate into vegetative cells and form biofilms that produce several substances including toxins and enzymes [7,8]. The biofilms allow B. cereus to survive in harsh environments. Moreover, the ability to form biofilms in bacteria allows the bacteria to resist antimicrobial agents and disinfectants, as well as to evade the host immune system [9,10].
The B. cereus group, also known as B. cereus sensu lato is composed of at least 11 species, including B. cereus sensu stricto (B. cereus s. s.), B. anthracis, B. thuringiensis, B. mycoides, B. pseudomycoides, B. weihenstephanensis, B. cytotoxicus, B. toyonensis, B. gaemokensis, B. manliponensis, and B. bingmayongensis [11]. Since the physiological-biological properties and the 16S rRNA sequences of the B. cereus group are similar, species of the B. cereus group may not be confirmed through traditional detection methods and phenotyping technologies [12]. Through next-generation sequencing technologies, whole bacterial genomes were evaluated, thereby enabling the establishment of moderate to high conservation of bacterial species via numerous proteins and aid in phylogeny identification [13,14].
Whole genome sequencing (WGS) of bacterial pathogens provides genome sequence information of the bacteria of interest, as well as of unknown bacteria, which can aid in revealing phylogenic and evolutionary trends [13,14]. The study of evolutionary trends and bacterial pathogenesis is beneficial for the development of novel antimicrobial agents or new strategies for pathogen control. Through complete genome sequencing of Bacillus halotolerans, tolerance potential to drought and salt stress was revealed [15]. Comparative genome analysis of Bacillus sporothermodurans allowed for characterization of the genes involved in heat resistance [16]. Key genes with insect resistance functions in Bacillus thuringiensis were characterized by complete genome sequencing, thereby providing genetic information for the development of potential bioinsecticides [17]. Some Bacillus sp. possess diarrheal toxin genes and virulence genes, which can be harmful to human health. Complete genome sequencing can be used to identify toxin and virulence genes, as well as genes related to biofilm formation in Bacillus pacificus isolate from food [18].
In this study, molecular approaches are used to characterize Bacillus spp. isolates from food samples with a heat resistance phenotype to identify evolutionary associations as well as to understand their heat resistance mechanisms. WGS analyses were performed in Bacillus sp. B48 and Bacillus sp. B140, isolates from an omelet with acacia leaves and fried rice, respectively [19]. Their closest phylogenetic neighbor and the genes involved in heat resistance were characterized to better understand this phenotype in B. cereus group. Moreover, molecular comparisons of these two isolates with mesophilic strain B. cereus ATCC 14579 and thermophilic strain B. cytotoxicus NVH 391-98 were also performed. This work provides evidence for the potential genetic basis for adaptation to heat conditions in the Bacillus species.
Bacillus sp. B48 and Bacillus sp. B140 isolates were isolated from an omelet with acacia leaves and fried rice [19], respectively. Glycerol stocks of the bacteria, stored at –80ºC, were streaked on nutrient agar plates and incubated overnight at 35 ± 2ºC.
The thermotolerance of B. cereus isolates was determined at a lethal temperature (60°C). The protocol was performed according to Periago et al. [20] with some modifications. Cultures in the mid-exponential growth phase (optical density at 600 nm of 0.5), were exposed to heat treatment at 60°C for 0, 1, 3, 6, 24, and 48 h. Viable counts were monitored by plate counting. Three independent experiments were performed for each isolate.
The antimicrobial susceptibility of Bacillus sp. B48 and Bacillus sp. B140 was determined using the Kirby–Bauer disk diffusion method according to standard criteria of the Clinical and Laboratory Standards Institute (CLSI) 2010 [21]. The antimicrobial discs tested in this study were procured from Himedia, Mumbai, India. The antimicrobial agents included ampicillin (AMP, 10 µg), amoxicillin-clavulanic acid (AMC, 20 µg/10 µg), penicillin G (PEN, 10 U), gentamicin (GEN, 10 µg), imipenem (IPM, 10 µg), vancomycin (VAN, 30 µg), chloramphenicol (CHL, 30 µg), ciprofloxacin (CIP, 5 µg), tetracycline (TET, 30 µg), and erythromycin (ERY, 15 µg). The isolates were classified as sensitive (S), intermediate (I), or resistant (R) to each antimicrobial agent based on the zones of inhibition, according to the interpretative criteria for Staphylococcus spp., following CLSI guidelines [22]. Staphylococcus aureus ATCC 25923 was assigned as a control strain for these antimicrobial susceptibility tests.
Genomic DNA (gDNA) was extracted and purified using a GF-1 Bacterial DNA Extraction Kit (Vivantis Technologies, Selangor, Malaysia) according to the manufacturer’s recommended protocol. The quality of the gDNA was determined by agarose gel electrophoresis and spectrophotometry. The gDNA libraries were prepared using a QIAGEN FX kit (Qiagen, Valencia, CA, USA). The quality and quantity of the indexed libraries were verified using an QIAxcel advance and a Denovix fluorometer. Then, the libraries were pooled in equimolar quantities and sequenced on an Illumina Miseq 2X250 bp paired-end (Illumina Inc., San Diego, CA, USA). Raw read quality was determined using the FASTQC software. Adaptors and poor-quality reads were removed using Trimmomatic, and the filtered reads were used as inputs for the Unicycler genome assembly program with default parameters [23].
The genomes of Bacillus sp. B48 and B140 were annotated using the Rapid Annotation using Subsystem Technology tool kit (RASTtk) in PATRIC (Pathosystems Resource Integration Center) [24]. Graphical circular genome maps were constructed by using the circular viewer in PATRIC. The virulence factors and antimicrobial resistance were also identified using PATRIC. The predicted coding sequence (CDS) was annotated from clusters of orthologous groups of proteins (COG) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using the alignment tool, BlastKOALA (https://www.kegg.jp/blastkoala/) and eggNOG-mapper. A Venn diagram was used to compare the CDS of Bacillus sp. B48 and Bacillus sp. B140 with those of B. cereus ATCC14579 and B. cytotoxicus NVH 391-98. The whole genome sequences of Bacillus sp. B48 and Bacillus sp. B140 were compared with that of B. cereus ATCC14579 and B. cytotoxicus NVH 391-98 using Mauve 2.3.1. A WGS-based phylogenetic tree was constructed using the CodonTree method within PATRIC webserver, which used the BV-BRC global Protein Families (PGFams) as homology groups [25]. The aligned protein and coding DNA were used to build the main tree with the RAxML analysis. Support values for the phylogenetic tree were generated using 100 rounds of the rapid bootstrapping option of RAxML [26]. FigTree (http://tree.bio.ed.ac.uk/software/figtree/) and iTOL were used for tree visualization [27]. Sequences of prophage Clp protease-like proteins from Bacillus sp. B48 and B140 were aligned using the Clustal Omega tool (https://www.ebi.ac.uk/Tools/msa/clustalo/).
The draft genomes of Bacillus sp. B48 and B140 were deposited in the NCBI SRA under the accession numbers SRR23924657 and SRR23924656, respectively. The BioSample accession for Bacillus sp. B48 and B140 were SAMN33833737 and SAMN33833738, respectively. The BioProject ID in GenBank is PRJNA946912.
The thermotolerance of B. cereus isolates was observed at a lethal temperature (60ºC) compared to mesophilic strain B. cereus ATCC 14579 as the control (Fig. 1). The colony counts of B. cereus ATCC14579 were dramatically decreased upon exposure to heat at 60ºC for 1 h and colonies were undetectable after 3 h. The colony counts of Bacillus sp. B48 and Bacillus sp. B140 decreased from log10 = 8 CFU/mL to approximately 2 after heat treatment for 1 h, and the constant colony count was observed even up to 48 h.
Bacillus sp. B48 and Bacillus sp. B140 were assessed for antimicrobial resistance to 10 antimicrobial agents (Table 1). Both isolates were susceptible to most of the tested antimicrobial agents, including GEN (26.6 ± 0.7 mm for Bacillus sp. B48 and 26.4 ± 0.4 mm for Bacillus sp. B140), IPM (37.3 ± 0.7 mm for Bacillus sp. B48 and 23.8 ± 1.9 mm for Bacillus sp. B140), VAN (23.1 ± 1.2 mm for Bacillus sp. B48 and 22.7 ± 0.6 mm for Bacillus sp. B140), CHL (31.5 ± 0.9 mm for Bacillus sp. B48 and 25.2 ± 0.7 mm for Bacillus sp. B140), CIP (32.0 ± 1.3 mm for Bacillus sp. B48 and 31.7 ± 0.6 mm for Bacillus sp. B140), TET (27.6 ± 1.1 mm for Bacillus sp. B48 and 27.5 ± 0.5 mm for Bacillus sp. B140), and ERY (29.2 ± 0.7 mm for Bacillus sp. B48 and 29.1 ± 0.9 mm for Bacillus sp. B140). However, both isolates were resistant to β-lactam antibiotics including AMP (10.3 ± 0.4 mm for Bacillus sp. B48 and 9.5 ± 0.6 mm for Bacillus sp. B140), AMC (5.9 ± 0.1 mm for Bacillus sp. B48 and 6.1 ± 0.2 mm for Bacillus sp. B140), and PEN (7.1 ± 0.5 mm for Bacillus sp. B48 and 7.8 ± 1.7 mm for Bacillus sp. B140).
The genomic features and annotation information are summarized in Table 2. The sequences of the genome drafts of Bacillus sp. B48 and Bacillus sp. B140 had estimated length of 5,533,408 bp and 5,279,040 bp with GC contents of 34.91% and 35.01%, respectively. The coding sequences were 5,705 bp for Bacillus sp. B48 and 5,468 bp for Bacillus sp. B140. The circular representation of the Bacillus sp. B48 and Bacillus sp. B140 draft genomes are shown in Fig. 2. The specialty genes of Bacillus sp. B48 and B140 including those involved in virulence factors and antibiotic resistance are summarized in Table 3.
The genomes of Bacillus sp. B48 and Bacillus sp. Bacillus sp. B140 were characterized using eggNOG database to analyze the COGs of proteins with functional annotations (Fig. 3). The majority of the annotated genes were classified as “Amino acid transport and metabolism” (E category), “General function prediction only” (R category), and “Transcription” (K category). These categories were similarly distributed in both strains. Functional annotation of both bacterial genomes was performed by the BlastKOALA tool on the KEGG Orthology (KO) database. A number of genes were classified as genetic information processing, signaling and cellular processing, environmental information processing, carbohydrate metabolism, and amino acid metabolism which being the most represented categories (Fig. 4).
The phylogenetic relationship of Bacillus species is illustrated in Fig. 5. Bacillus sp. B48 was found to be closely related to B. cereus while Bacillus sp. B140 was found to be closely related to B. thuringiensis.
Analysis of orthologous gene clusters using OrthoVenn2 demonstrated that a core of 3,058 orthologous genes was shared among the genomes of Bacillus sp. B48, Bacillus sp. B140, B. cereus ATCC14579, and B. cytotoxicus NVH 391-98 (Fig. 6). There were 506, 339, 382, and 541 singleton gene clusters present in Bacillus sp. B48, Bacillus sp. B140, B. cereus ATCC14579, and B. cytotoxicus NVH 391-98, respectively (Table 4). The genomes of Bacillus sp. B48, Bacillus sp. B140, B. cereus ATCC14579, and B. cytotoxicus NVH 391-98 contained 21, 9, 8, and 21 unique genes, respectively (Supplementary Table 1).
Whole genome alignments of Bacillus sp. B48, Bacillus sp. B140, B. cereus ATCC14579, and B. cytotoxicus NVH 391-98 revealed the existence of numerous homologous regions (Fig. 7). The genomic arrangement of the heat shock protein grpE, as well as associated genes responsible for heat shock in the dnaK gene cluster, are shown in Fig. 8. The similarity of chromosomal regions of the heat shock proteins was observed in all four strains except for the presence of DUF4253 domain-containing protein in the chromosome of B. cereus ATCC14579 and the intracellular protease, PfpI family, in the chromosome of B. cytotoxicus NVH 391-98.
PATRIC analysis showed that the prophage Clp protease-like proteins were present in the genome of Bacillus sp. B48 and Bacillus sp. B140. Fig. 9 shows the amino acid sequence alignments of the prophage Clp protease-like proteins of Bacillus sp. B48 and Bacillus sp. B140 with low similarity.
In this study, we genetically characterized food-isolated strains of Bacillus sp. B48 and Bacillus sp. B140. Both strains are resistant to heat treatment at 60°C for at least 48 h. Therefore, it is interesting to study their genomic profiles, especially those related to heat responses such as heat shock gene clusters. Several emerging highly heat-resistant Bacillus species have been previously classified including B. thermoamylovorans, B. sporothermodurans, and B. cytotoxicus NVH 391-98 [16,28,29]. B. sporothermodurans can significantly affect the quality of heat-processed foods since it is able to form heat-resistant spores that can survive in foods treated with ultra-high temperatures [16]. B. thermoamylovorans is an emerging highly heat-resistant Bacillus species isolated from milk [28]. B. cytotoxicus NVH 391-98, a thermotolerant species of the B. cereus group, was isolated during a severe food poisoning outbreak in France in 1998 [29]. However, phylogenetic analysis suggested that both Bacillus sp. B48 and Bacillus sp. B140 are not closely related to B. thermoamylovorans, B. sporothermodurans, or B. cytotoxicus NVH 391-98. Based on whole genome phylogenetic analysis, Bacillus sp. B48 and Bacillus sp. B140 were found to be closely related to B. cereus and B. thuringiensis, respectively. Therefore, the categorization of bacterial inhabitants is based on sequence similarity threshold rather than biological relevance [16,30].
Heat shock proteins (HSPs) are responsible for heat shock treatment including thermal food processing. Several conditions other than heat shock can induce HSP expression, these include cold, ultraviolet light, and during cell healing [31]. Major HSPs that have been reported in prokaryotes include GroES, GrpE, DnaJ, GroEL, DnaK, HtpG, ClpB, ClpA, and ClpX [31-35]. Most of these HSPs are present in B. cereus ATCC14579 and B. cytotoxicus NVH 391-98, as well as in Bacillus sp. B48 and Bacillus sp. B140. These molecular chaperones are important because they prevent protein aggregation during heat stress [16,20]. The crucial process of high heat resistance in Bacillus spp. during periods of stress and sporulation requires ClpA, ClpC, ClpE, ClpP, ClpQ, ClpX, and ClpY [36]. In various prokaryotes, the ClpB, ClpA, and ClpX protein complex modulates the tolerance to extreme temperatures by remodeling and degrading aggregated proteins [37]. ClpB, which was identified in B. subtilis, B. sporothermodurans, and B. oleronius, works together with the chaperones DnaJ, DnaK, and GrpE to repair heat-induced protein damage [38]. In B. subtilis, ClpX together with ClpP play a role in cell survival at thermal stress under limiting levels of the DnaK system [39]. We found that both Bacillus sp. B48 and Bacillus sp. B140, but not B. cereus ATCC14579, contain prophage Clp protease-like proteins. These proteins may be involved in the heat-tolerant phenotype in both isolates. However, it is necessary to further characterize the role of these proteins especially relating to the heat-resistant phenotype.
HrcA is a heat-inducible transcription repressor detected in B. subtilis and B. sporothermodurans. DnaK, DnaJ, and GrpE are involved in the repair of heat-induced protein damage by forming a cellular chaperone machinery [33]. GrpE is the chaperone moiety of the dnaK operon that works together with GroEL for the renaturation of heat-denatured proteins [33]. Many of these proteins are shared amongst the Bacillus species and they are also homologs of each other, with some variations in the amino acid sequences. All four strains mentioned in this study also possess these proteins with some variations in amino acid sequences. However, the DUF4253 domain-containing protein is present only in B. cereus ATCC14579 but not in Bacillus sp. B48, Bacillus sp. B140 or B. cytotoxicus NVH 391-98. Four domains of unknown function including DUF438, DUF1524, DUF1957, and DUF3458_C, have been hypothesized to play roles in temperature adaptation in both archaea and bacteria [40]. Therefore, the function of this protein in heat response in Bacillus species needs to be further characterized.
In conclusion, B. cereus, a foodborne pathogen that can cause diarrheal or emetic food poisoning, can grow and survive under varied temperatures with diverse heat-responsive mechanisms, especially those that require HSPs. In this study, a genomic analysis of food-isolated Bacillus sp. B48 and Bacillus sp. B140 with thermotolerant phenotype revealed the presence of prophage Clp protease-like proteins with low amino acid sequences similarities. This protein was not present in the genome of the mesophilic strain, B. cereus ATCC14579. Further experimental analysis in terms of gene knockout and protein characterization is required to confirm the function of prophage Clp protease-like proteins relating to heat tolerant phenotype of the bacteria, especially in Bacillus species.
Notes
Acknowledgments
This research was financially supported by the Faculty of Medicine, Thammasat University, Thailand. The grant number is 2-04/2565. This work was also supported by the Research Group in Multidrug Resistant Bacteria and the Antimicrobial Herbal Extracts, Faculty of Medicine, Thammasat University, and Thammasat University Research Unit in Nutraceuticals and Food Safety.
Supplementary Materials
Supplementary data can be found with this article online at http://www.genominfo.org.
Supplementary Table 1.
Cluster list of the unique genes observed in Bacillus sp. B48 and Bacillus sp. B104 genomes using OrthoVenn2
Fig. 1.
Thermotolerance of Bacillus sp. B48 and Bacillus sp. B140. Log-phase bacterial cells were exposed to heat at 60 ºC for 0, 1, 3, 6, 24, and 48 h. Colony counts were performed after incubation on nutrient agar plates at 35 ± 2ºC overnight. Three independent experiments were performed for each isolate. B48, Bacillus sp. B48; B140, Bacillus sp. B140; ATCC14579, B. cereus ATCC14579.
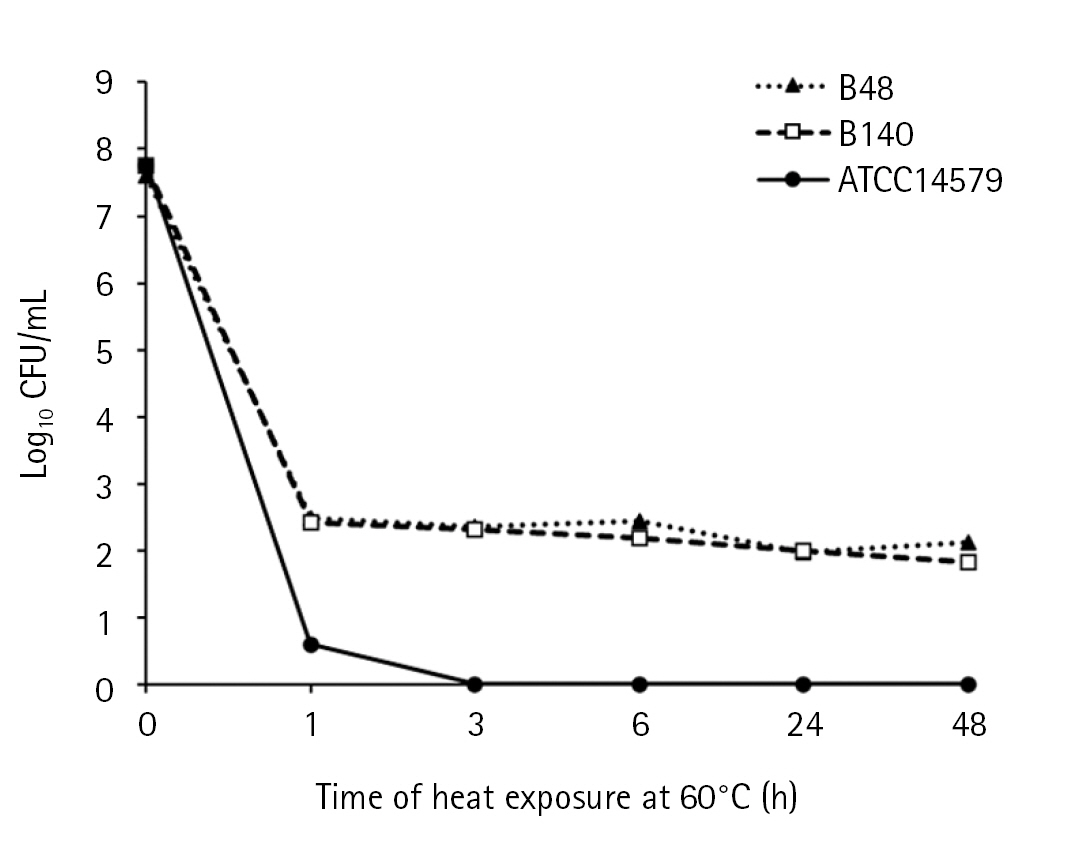
Fig. 2.
Genomic circular diagram of Bacillus sp. B48 and Bacillus sp. B140. A genome circular diagram was reconstructed using the circular viewer of PATRIC, from the outer circle to the inner circle: contigs, forward coding sequence (CDS), reverse CDS, non-CDS features, AMR genes, VF genes, transporters, and drug targets. The two inner tracks are GC content and GC skew.
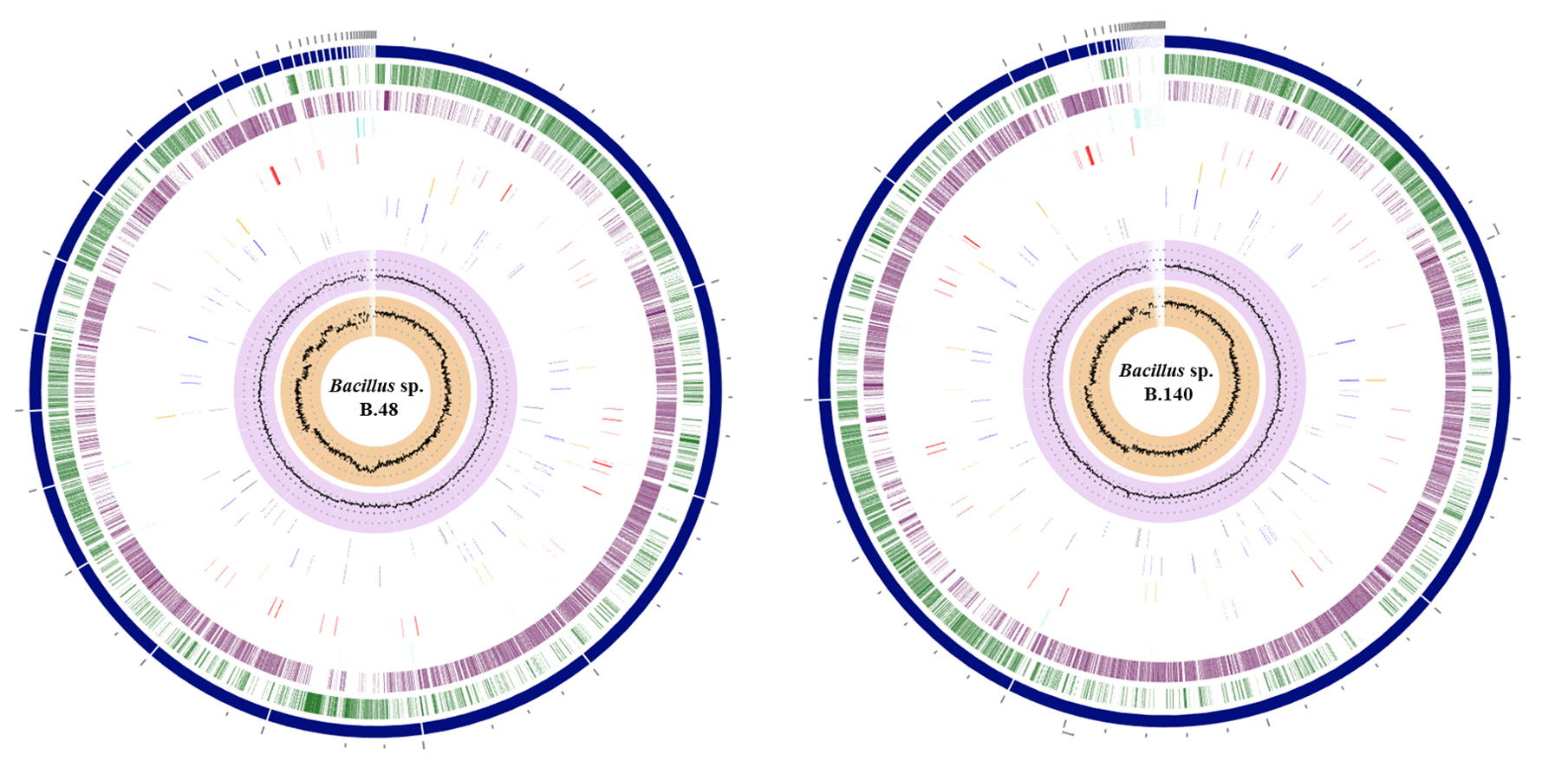
Fig. 3.
The clusters of orthologous groups of proteins (COG) function annotation of Bacillus sp. B48 (B48) and Bacillus sp. B140 (B140). The X-axis represents the functional classification of COG. The Y-axis represents the number of genes annotated in each classification. The details of each classification are illustrated on the right.
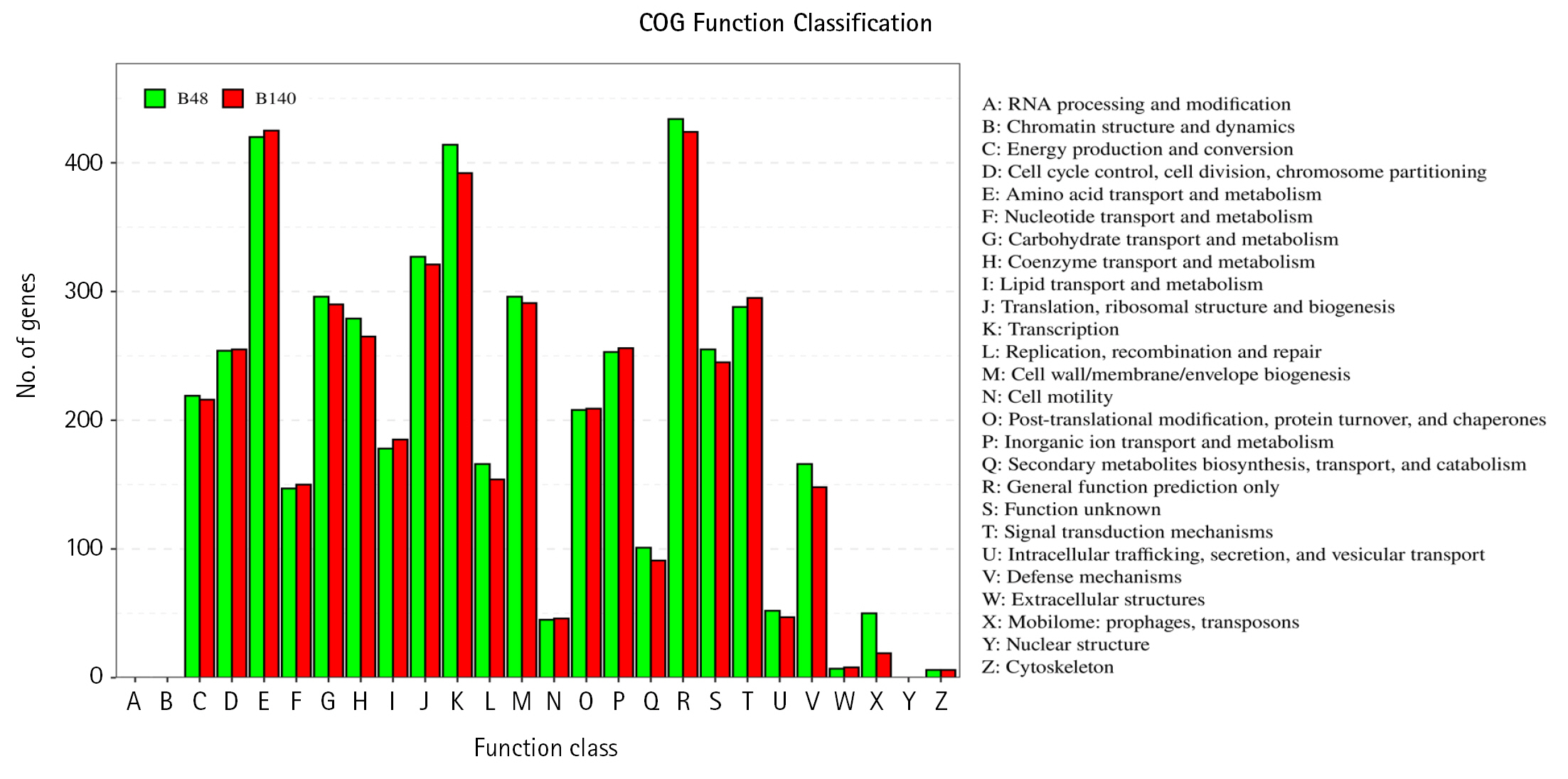
Fig. 4.
Genome annotation of Bacillus sp. B48 and Bacillus sp. B140 by BlastKOALA Kyoto Encyclopedia of Genes and Genomes (KEGG). The total number of proteins assigned to the most abundant categories are illustrated.

Fig. 5.
Whole genome sequencing–based phylogenetic tree of Bacillus sp. B48 and Bacillus sp. B140. The codon tree method selects single-copy PATRIC PGFams. Aligned proteins and coding DNA from single-copy genes were analyzed using the RAxML program. FigTree and iTOL were used for tree visualization. Escherichia coli was used as the outgroup.
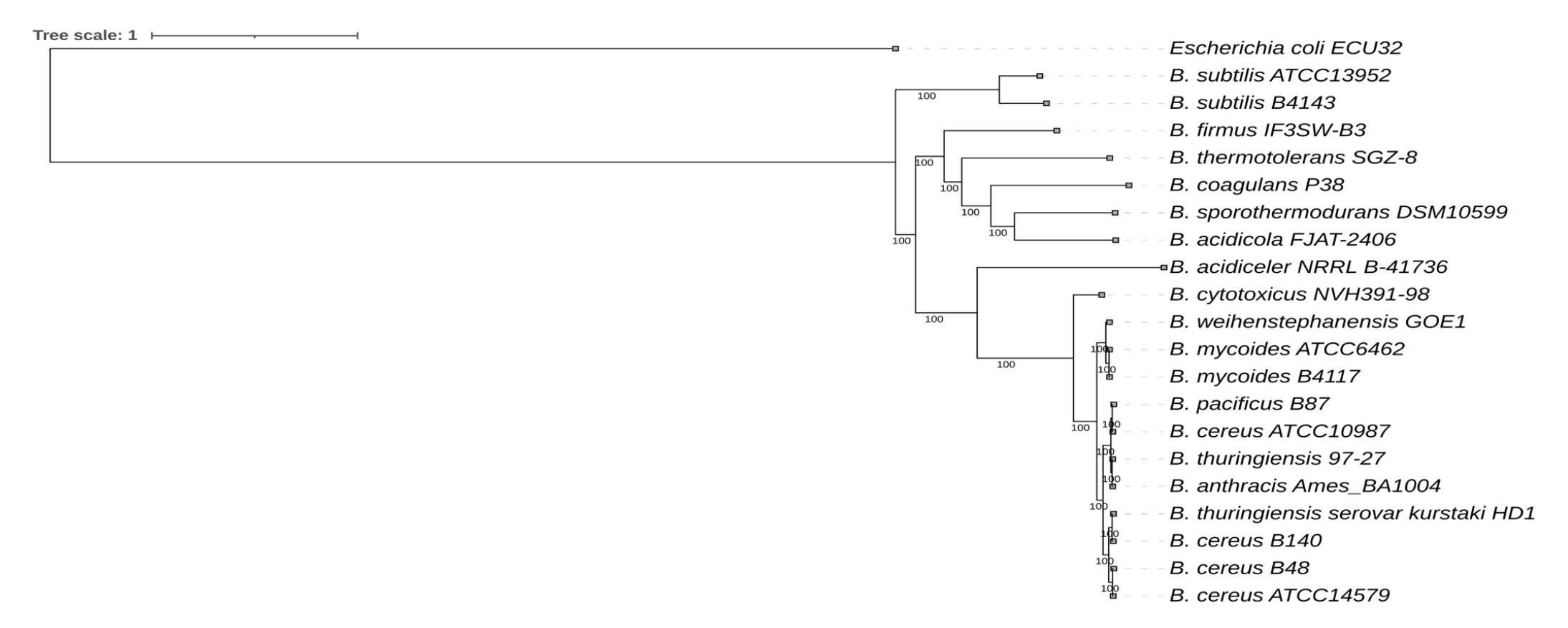
Fig. 6.
Venn diagram of Bacillus sp. B48, Bacillus sp. B140, B. cereus ATCC14579, and B. cytotoxicus NVH 391-98. Whole genome comparison and annotation of orthologous gene clusters were performed using OrthoVenn2.

Fig. 7.
Whole genome alignment of Bacillus sp. B48, Bacillus sp. B140, B. cereus ATCC14579, and B. cytotoxicus NVH 391-98 using the Mauve program. Strain names are indicated on the left. Regions with homologous sequences are illustrated with the same-colored squares.

Fig. 8.
Comparison of chromosomal regions of the heat shock protein GrpE of Bacillus sp. B48, Bacillus sp. B140, B. cereus ATCC14579, and B. cytotoxicus NVH 391-98 in the Mauve program. The details of each number are described on the right.
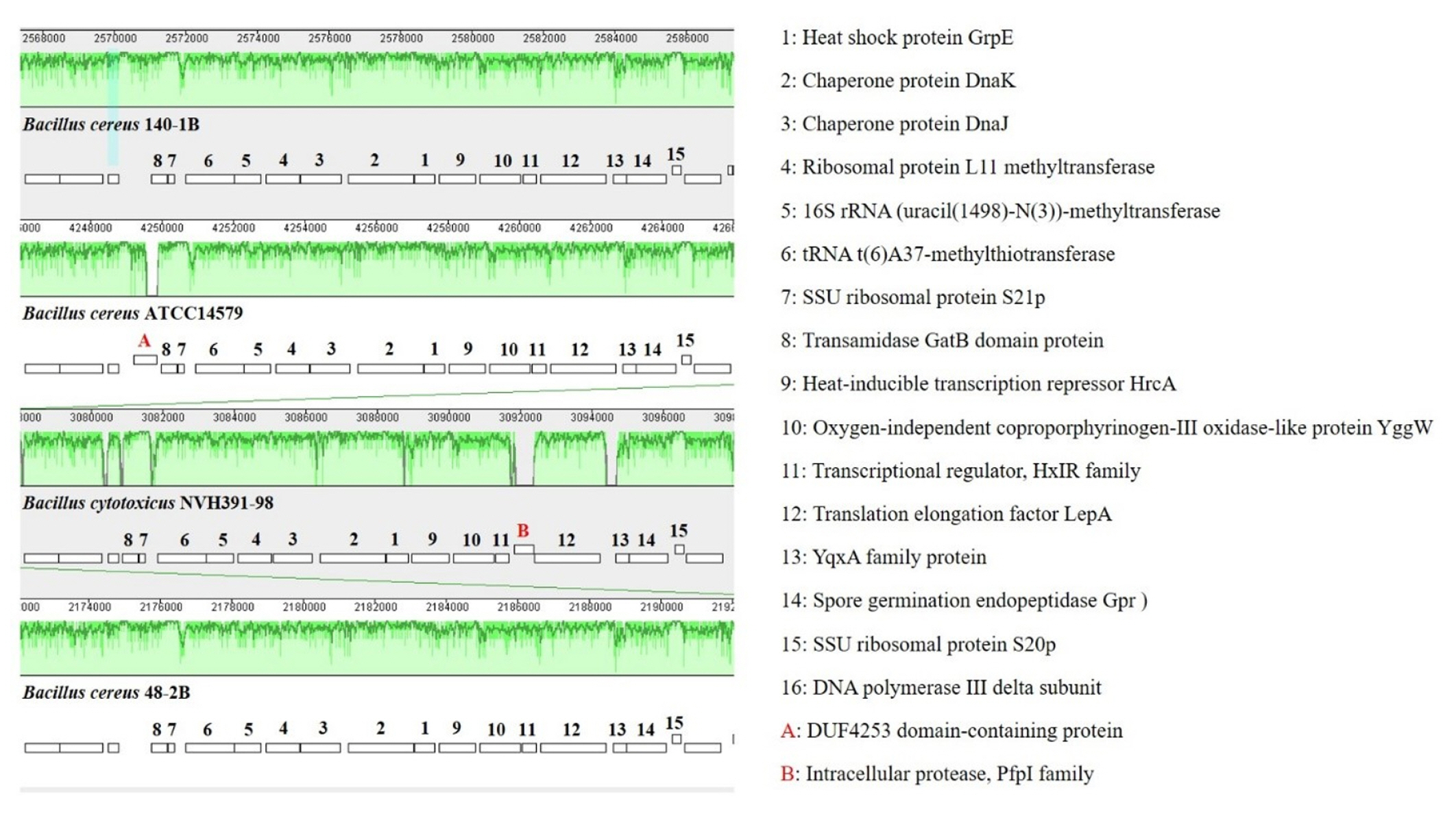
Fig. 9.
Amino acid sequence alignment of the prophage Clp protease-like proteins of Bacillus sp. B48 (B48) and Bacillus sp. B140 (B140). Sequences alignments were performed using Clustal Omega.

Table 1.
Antimicrobial susceptibility testing of Bacillus sp. B48 and Bacillus sp. B140
Table 2.
Genomic features and annotation information of the chromosome of Bacillus sp. B48 and Bacillus sp. B140
| Genome feature | Bacillus sp. B48 | Bacillus sp. B140 |
|---|---|---|
| Genome length (bp) | 5,533,408 | 5,279,040 |
| Protein-coding genes | 5,705 | 5,468 |
| GC content (%) | 34.91 | 35.01 |
| No. of tRNAs | 54 | 71 |
| No. of rRNAs | 4 | 6 |
| Contigs | 38 | 35 |
Table 3.
Specialty genes in genomic sequence of Bacillus sp. B48 and Bacillus sp. B140 annotated using Subsystem Technology tool kit (RASTtk) in PATRIC
References
1. Wijnands LM, Dufrenne JB, Zwietering MH, van Leusden FM. Spores from mesophilic Bacillus cereus strains germinate better and grow faster in simulated gastro-intestinal conditions than spores from psychrotrophic strains. Int J Food Microbiol 2006;112:120–128.


2. Altayar M, Sutherland AD. Bacillus cereus is common in the environment but emetic toxin producing isolates are rare. J Appl Microbiol 2006;100:7–14.


3. Owusu-Kwarteng J, Wuni A, Akabanda F, Tano-Debrah K, Jespersen L. Prevalence, virulence factor genes and antibiotic resistance of Bacillus cereus sensu lato isolated from dairy farms and traditional dairy products. BMC Microbiol 2017;17:65.




4. Webb MD, Barker GC, Goodburn KE, Peck MW. Risk presented to minimally processed chilled foods by psychrotrophic Bacillus cereus. Trends Food Sci Technol 2019;93:94–105.



5. Auger S, Galleron N, Bidnenko E, Ehrlich SD, Lapidus A, Sorokin A. The genetically remote pathogenic strain NVH391-98 of the Bacillus cereus group is representative of a cluster of thermophilic strains. Appl Environ Microbiol 2008;74:1276–1280.



6. Kumari S, Sarkar PK. Bacillus cereus hazard and control in industrial dairy processing environment. Food Control 2016;69:20–29.

7. Scheldeman P, Herman L, Foster S, Heyndrickx M. Bacillus sporothermodurans and other highly heat-resistant spore formers in milk. J Appl Microbiol 2006;101:542–555.


8. Esteban MD, Huertas JP, Fernandez PS, Palop A. Effect of the medium characteristics and the heating and cooling rates on the nonisothermal heat resistance of Bacillus sporothermodurans IC4 spores. Food Microbiol 2013;34:158–163.


9. Caro-Astorga J, Frenzel E, Perkins JR, Alvarez-Mena A, de Vicente A, Ranea JA, et al. Biofilm formation displays intrinsic offensive and defensive features of Bacillus cereus. NPJ Biofilms Microbiomes 2020;6:3.




10. Ryu JH, Beuchat LR. Biofilm formation and sporulation by Bacillus cereus on a stainless steel surface and subsequent resistance of vegetative cells and spores to chlorine, chlorine dioxide, and a peroxyacetic acid-based sanitizer. J Food Prot 2005;68:2614–2622.



11. Liu Y, Lai Q, Du J, Shao Z. Genetic diversity and population structure of the Bacillus cereus group bacteria from diverse marine environments. Sci Rep 2017;7:689.




12. Liu Y, Ge W, Zhang J, Li X, Wu X, Li T, et al. Detection of Bacillus cereus sensu lato from environments associated with goat milk powdered infant formula production facilities. Int Dairy J 2018;83:10–16.

13. Alcaraz LD, Moreno-Hagelsieb G, Eguiarte LE, Souza V, Herrera-Estrella L, Olmedo G. Understanding the evolutionary relationships and major traits of Bacillus through comparative genomics. BMC Genomics 2010;11:332.




14. Zwick ME, Joseph SJ, Didelot X, Chen PE, Bishop-Lilly KA, Stewart AC, et al. Genomic characterization of the Bacillus cereus sensu lato species: backdrop to the evolution of Bacillus anthracis. Genome Res 2012;22:1512–1524.



15. Zhang Z, Yin L, Li X, Zhang C, Liu C, Wu Z. The complete genome sequence of Bacillus halotolerans ZB201702 isolated from a drought- and salt-stressed rhizosphere soil. Microb Pathog 2018;123:246–249.


16. Owusu-Darko R, Allam M, Ismail A, Ferreira CAS, Oliveira SD, Buys EM. Comparative genome analysis of Bacillus sporothermodurans with its closest phylogenetic neighbor, Bacillus oleronius, and Bacillus cereus and Bacillus subtilis groups. Microorganisms 2020;8:1185.



17. Zribi Zghal R, Ghedira K, Elleuch J, Kharrat M, Tounsi S. Genome sequence analysis of a novel Bacillus thuringiensis strain BLB406 active against Aedes aegypti larvae, a novel potential bioinsecticide. Int J Biol Macromol 2018;116:1153–1162.


18. Sornchuer P, Saninjuk K, Prathaphan P, Tiengtip R, Wattanaphansak S. Antimicrobial susceptibility profile and whole-genome analysis of a strong biofilm-forming Bacillus sp. B87 strain isolated from food. Microorganisms 2022;10:252.



19. Sornchuer P, Tiengtip R. Prevalence, virulence genes, and antimicrobial resistance of Bacillus cereus isolated from foodstuffs in Pathum Thani Province, Thailand. Pharm Sci Asia 2021;48:194–203.

20. Periago PM, van Schaik W, Abee T, Wouters JA. Identification of proteins involved in the heat stress response of Bacillus cereus ATCC 14579. Appl Environ Microbiol 2002;68:3486–3495.




21. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 20th informational supplement. CLSI document M100-S20. Wayne, PA: Clinical and Laboratory Standards Institute, 2010.
22. Gao T, Ding Y, Wu Q, Wang J, Zhang J, Yu S, et al. Prevalence, virulence genes, antimicrobial susceptibility, and genetic diversity of Bacillus cereus isolated from pasteurized milk in China. Front Microbiol 2018;9:533.



23. Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017;13:e1005595.



24. Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 2015;5:8365.




25. Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res 2017;45:D535–D542.


26. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30:1312–1313.




27. Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 2007;23:127–128.



28. Flint S, Gonzaga ZJ, Good J, Palmer J. Bacillus thermoamylovorans: a new threat to the dairy industry. A review. Int Dairy J 2017;65:38–43.

29. Guinebretiere MH, Auger S, Galleron N, Contzen M, De Sarrau B, De Buyser ML, et al. Bacillus cytotoxicus sp. nov. is a novel thermotolerant species of the Bacillus cereus group occasionally associated with food poisoning. Int J Syst Evol Microbiol 2013;63:31–40.


31. Hecker M, Schumann W, Volker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol 1996;19:417–428.



32. Liberek K, Georgopoulos C. Autoregulation of the Escherichia coli heat shock response by the DnaK and DnaJ heat shock proteins. Proc Natl Acad Sci U S A 1993;90:11019–11023.



33. Schroder H, Langer T, Hartl FU, Bukau B. DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J 1993;12:4137–4144.




34. Gamer J, Multhaup G, Tomoyasu T, McCarty JS, Rudiger S, Schonfeld HJ, et al. A cycle of binding and release of the DnaK, DnaJ and GrpE chaperones regulates activity of the Escherichia coli heat shock transcription factor sigma32. EMBO J 1996;15:607–617.




36. Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 1998;94:73–82.


37. LaBreck CJ, May S, Viola MG, Conti J, Camberg JL. The protein chaperone ClpX targets native and non-native aggregated substrates for remodeling, disassembly, and degradation with ClpP. Front Mol Biosci 2017;4:26.



38. Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol Microbiol 2001;40:397–413.


- TOOLS
- Related articles in GNI








