|
 |
- Search
| Genomics Inform > Volume 20(1); 2022 > Article |
|
Abstract
Human exposure to pollutants has been on the rise. Thus, researchers have been focused on understanding the effect of these compounds on human health, especially on the genetic information by using various tests, among them the somatic mutation and recombination tests (SMARTs). It is a sensitive and accurate method applicable to genotoxicity analysis. Here, a comprehensive bibliometric analysis of SMART assays in genotoxicity studies was performed to assess publication trends of this field. Data were extracted from the Web of Science database and analyzed by the bibliometric tools HistCite, Biblioshiny (RStudio), VOSViewer, and CiteSpace. Results have shown an increase in the last 10 years in terms of publication. A total of 392 records were published in 96 sources mainly from Brazil, Spain, and Turkey. Research collaboration networks between countries and authors were performed. Based on document co-citation, five large research clusters were identified and analyzed. The youngest research frontier emphasized on nanoparticles. With this study, how research trends evolve over years was demonstrated. Thus, international collaboration could be enhanced, and a promising field could be developed.
Anthropogenic activities disseminate large amounts of chemical substances into the environment [1]. Hence, humans are currently exposed to several pollutants with genotoxic potentials such as metals [2], pesticides [3], industrial waste mix [4], and nanomaterials [5]. Genotoxicity is a wide term comprising DNA damage and mutagenicity, where the mutagenic effect is described as an occurred event with irreversible and heritable outcomes affecting the DNA and/or chromosome structure [6]. The genotoxic effect recorded on somatic cells has been associated with pathological endpoints such as premature aging, neuronal diseases, and even carcinogenesis [7,8]. Accordingly, the genotoxicity evaluation is a required component in the human health risk assessment and as one single test is unable to detect all the genotoxic endpoints, a battery of in vitro and in vivo tests was recommended [9]. Among these assays, the somatic mutation and recombination tests (SMARTs) are one of the commonly used tests. The SMART assays are in vivo assay to assess the potential genotoxicity of substances in the somatic cells of Drosophila melanogaster [10]. This assay could target wing cells named as wing-spot test proposed first time by Graf et al. [11], or eyes cells known as eye-spot test defined by Wurgler and Vogel [12]. In both cases, losing heterozygosity by deletions, point mutations, mitotic recombination, and nondisjunction unravels the expression of genetic markers in heterozygous or transheterozygous individuals, ensuring the quantification of the damage by visual scoring [13]. The wing-spot test includes two types of cross using recessive genetic markers on the 3rd chromosome, a standard cross with normal bioactivation between female virgins (flr3/In (3LR)TM3, ri ppsep l(3)89Aa bx34e e Bds) and (mwh/mwh) males, and a high metabolic bioactivation cross with high levels of cytochrome P450 between female virgins (ORR; flr3/In (3LR)TM3, ri ppsep l(3)89Aa bx34e e Bds) and (mwh/mwh) males. The mutant spots are produced after chemical exposure that induced point mutation, deletion, or mitotic recombination [14,15]. The eye-spot test is based on a cross between wild-type eyed females (w+/w+) and white-eyed males (w/Y). The gene white (w) is a recessive marker found on the X chromosome. During the offspring period, a mutagenic event could occur and cause the formation of white phenotype spots (mutant ommatidia) in the wild-type eyes [16]. Although both tests are accurate, sensitive, and specific, the wing-spot test allows the visual scoring of wings over time, whereas in the eye-spot test, the analysis should be performed quickly since no preserving actions are available on the eyes [17]. In fact, the SMART assays have been applied in the analysis of the genotoxicity and the antimutagenicity of several chemicals and agents such as pesticides [18-20], nanomaterials [21,22], food products [23,24], hormones [25], plant extracts [26], and drugs [27-29]. With this variety of studies, a bibliometric analysis is required to assess the impact of this methodology on the genotoxicity studies. Bibliometric analysis is considered a highly sensitive method to evaluate research outputs based on statistical tools and to study metrological features of information created in a specific field [30]. To the best of our knowledge, no paper using the bibliometric analysis to explore the trends of SMART assays research has been published. Therefore, in the current study, various aspects were exanimated to evaluate the publications and citation trends in SMART assays from 1984 to 2020. Hence, the following research objectives were considered guiding the study design: (1) to identify the most influential journals and publications, the impactful authors and institutions, and the leading countries in SMARTs literature; (2) to find the patterns of collaboration between countries and authors within this research domain; (3) to explore the emerging keywords and research themes. The findings of the present study will provide a comprehensive overview of the importance of SMART assays, it would provide information to the scholars to easily identify the research profile and enhance collaboration.
This bibliometric study analyzed the published academic studies of SMARTs indexed in the Web of Science (WOS) core collection database. As known, WOS is the most reliable global citation database with a collection of over 21,000 peer-reviewed journals and the most accepted one for analysis of academic papers [31]. A comprehensive four-step approach was framed in this study as shown in Fig. 1. Boolean operators were used with adding all the relevant keywords to retrieve more relevant papers. (TS = ((“SMART assays” OR “SMART assay” OR “SMART test” OR “SMART tests” OR “wing-spot test” OR “wing-spot assay” OR “somatic mutation and recombination test” OR “eye-spot test” OR “eye-spot assay” OR “small single spots” OR “large single spots” OR “mutant ommatidia” OR “mutant eye unit” OR “w/w+ SMART”))). The search was performed on 14 January 2021; it is essential to present the date of records collection as the database is constantly updating [32]. Most bibliometric studies are based on academic articles [30], thereby this study was limited to original articles written in English in the strict sense. The search was also limited to 2020. The authors adopted the PRISMA approach, which has been used in bibliometric studies. A total of 460 records were extracted initially, which later were filtered by document types, to exclude the following document types: proceedings papers, book chapters, and early access. The data relevance and accuracy were assured by scanning the title and abstract of each record, and 49 records were excluded. The irrelevant articles were from the following categories: (1) Internet of things and wireless sensing, (2) communication services, (3) image processing, (4) industry, and (5) single molecule amplification and re-sequencing technology (SMART). Finally, a total of 392 records were remained to be downloaded in plain text to extract the following data: publication year, author, title, abstract, keywords, cited references, journal title, and institution, for further analysis.
Having selected 392 relevant records, data visualization and analysis were conducted by using various bibliometric software and applications, including Microsoft Excel, HistCite, Biblioshiny (RStudio), VOSViewer, and CiteSpace. The analysis was performed in three stages. First, a bibliometric citation analysis was performed (number of publications and citations, relevant journals, productive institutions, and the most impactful articles and authors). Secondly, a network analysis was applied, including collaboration between countries and co-authoring using the Walktrap clustering algorithm. This algorithm has the advantage to be computed effectively and placing the data in a network [33]. Furthermore, a three-fields-plot based on the Sankey diagram to indicate the interrelation between keywords, journals, and countries was visualized. Detection of the emerging research fields is required to outline research areas. Therefore, thirdly, a content analysis was conducted. HistCite (version 12.03.17) was used for the bibliometric citation analysis to sort the collected data by quantitative (number of publications and citations) and qualitative indicators (total global citation score and total local global citation score). Hence, these two types of indicators were applied in the current study. The total global citation score (TGCS) represents the number of citations of a paper included in the collection selected for the analysis in the WOS whereas the total local citation score (TLCS) refers to the total number of citations of a paper included in the collection and has been cited by other papers of the same collection [34]. The RStudio software (version 1.3.1093) with Biblioshiny application was used [35]. Hence, this application was also used to analyze the basic indicators of the search, plus the collaboration between countries and authors, and the three-fields plot. VOSViewer software (version 1.6.16) was used to analyze and visualize the emerging keywords. CiteSpace (version 5.7.R4) was used to perform the co-citation analysis and to investigate the emerging topics. Co-citation analysis is an effective tool to understand the intellectual structure of a research field, with the cited papers intellectual bases are revealed whereas papers in their active state of citation represent the research frontiers which usually display characteristics of a field specificity [36]. One of the important tools found in CiteSpace is the betweenness centrality. A paper with high betweenness centrality was defined as an important research paper in the network [37]. Furthermore, articles with strong citation bursts with time slices were identified. The burst is observed when a publication has an excess in its citation counts compared to its peers. This furthers identify publications that interested the scholars over time and thereby help to explore the research frontiers of a given field [38,39].
The present study analyzed the SMART assays in the genotoxicity studies published during 1984-2020. A total of 392 records have been written by 933 authors from 35 different countries with an average of 17.01 citations per document. Authors of single-authored documents are seven (0.75%) while authors of multi-authored documents represent 926 authors (99.25%).
The first publication appeared in 1984, thereafter the number of publications has been gradually increasing at a rate of 14.4 articles per year with a registered bloom in 2013 and 2015 (Recs, 22), whereas the TLCS and TGCS have recorded the bloom in 1984 (TLCS, 319; TGCS, 527) and in 1992 (TLCS, 282; TGCS, 547). The output of the first 25 years (1984‒2009) was 206 publications and only in the past 10 years (2010‒2020), 186 studies were published. This finding indicates the growing interest in using SMART assays in genotoxicity studies (Fig. 2).
The top 10 authors collectively contributed to 302 studies. Graf U, Marcos R, and de Andrade HHR are the top three authors in SMART assays field (Table 1). Graf U is the most influential author with the highest number of TLCS (953) and TGCS (1,716).
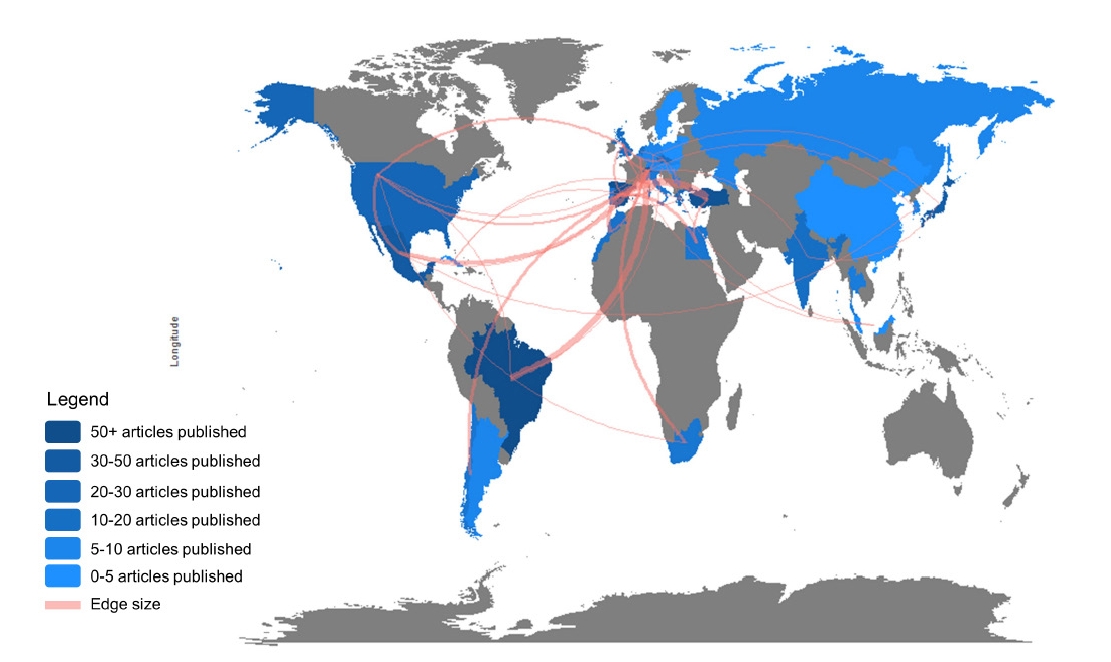
The top 10 institutions published 70.2% (n = 275) studies of the total publications. The Federal University of Uberlândia in Brazil is on the top of the list with 39 articles (TGCS, 425) (Table 2). Interestingly, the University of Zürich ranked in 4th position has the highest TGCS (1,892) and TLCS (1,119). There is only one country having a number of publications in three-digits, Brazil, with 106 publications and 1,103 TGCS, followed by Spain (TGCS, 1,470; TLCS, 338) and Turkey (TGCS, 784; TLCS, 221).
The 392 studies in SMART assays were published in 96 academic sources. More than half of these studies (n = 241, 61.5%) were published in the top 10 journals. The sources ‘Food and Chemical Toxicology’ and ‘Mutation Research Genetic Toxicology and Environmental Mutagenesis’ are on the top of the list with 53 publications (Table 3). The journal with the highest impact factor Chemosphere (7.086) has published 10 studies with 177 TGCS.
The years of the top 10 highly cited articles ranged from 1984 to 1996. There is only one article that obtained over 200 citations. This article has been titled ‘Somatic Mutation and Recombination Test in Drosophila melanogaster’ by Graf et al. and published in 1984 [11]. Half of the highly cited articles were published in Mutation Research journal, and this journal is on the top list of influential journals (Table 4).
Each node represents an author and the edges indicate the research collaboration between them. Six clusters are recorded, the blue and the orange clusters both with five authors are the largest clusters, followed by the purple cluster which includes four authors (Fig. 3). The brown cluster has one separate author.
A total of 38 collaboration entries are registered worldwide. Turkey, Brazil, and Mexico were the most collaborative countries (Fig. 4).
The co-occurrence of keywords represents the relationship between two words that occurred together. Three minimum number of occurrences of a keyword were selected; hence, out of 861 authors’ keywords, 70 meet this criterion to form 11 clusters (Fig. 5). Each color indicates a separate cluster and clusters are organized based on the link strength and occurrence. Thus, the size of the bubble represents the relationship between link strength and occurrence. The first five keywords with the high total link strength are Drosophila melanogaster (link strength: 320), genotoxicity (247), SMART (181), Drosophila (108), and wing-spot test (105).
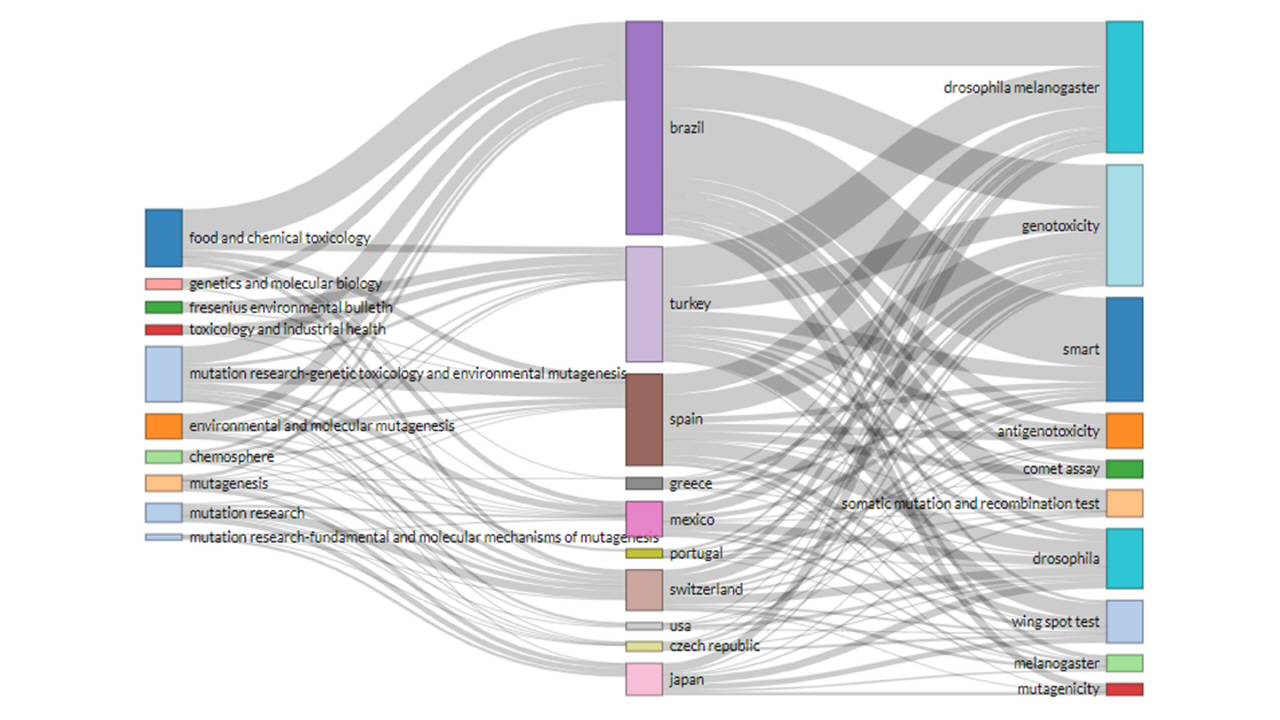
The interconnections among sources (left), countries (middle), and author keywords (right) are analyzed to understand which keywords are preferable to which countries and used to what sources. The three top countries (Brazil, Turkey, and Spain) have a strong connection with the source ‘Food and Chemical Toxicology’ and prefer to publish four keywords (Drosophila melanogaster, genotoxicity, SMART, and antigenotoxicity). The block length presented in Fig. 6 indicates the level of connection.
A co-citation network was generated with 985 nodes and 3,291 links for a one-year time slice (Fig. 7). The cited references are representing in the form of nodes and the co-citation relationships are visualized in the form of links. The top five co-cited articles are shown in Table 5 [14,41,47-49]. From a total of 392 records and 16,734 references, a list of the top 25 references with the strongest citation bursts was generated (Fig. 8). The citation bursts in the list of the top 25 references have been expanded between 1989 and 2017. Most of the strength bursts ranged between 4 and 7, and most citations have 3 to 4 years expanded duration. However, the citation with the most expanded duration (2011‒2016) has a low strength of its citation bursts (5.47) [50]. The reference paper written by Frei and Wurgler (1988) had the highest citation burst (16.12) [47].
A total of 17 clusters were identified in the SMART assays literature. The largest clusters are shown in Fig. 9. Based on specific metrics, term frequency‒inverse document frequency, log-likelihood tests (LLR), and mutual information tests, CiteSpace analyses the title of articles to extract a noun to characterize the cluster type. Generally, LLR covers the best themes (Table 6). A silhouette value (S) >0.7 denotes the high credibility of a cluster and a value of modularity (Q) >0.3 reveals the significant structure of the network [51]. As shown in Figs. 7 and 8, the top-ranked item by centrality is Frei and Wurgler (1995) [40] in Cluster #2, with the centrality of 41. The second one is Carmona et al. (2011) [50] in Cluster #3, with the centrality of 38. The third is Demir et al. (2011) [49] in Cluster #2, with the centrality of 34. Therefore, these papers are considered as pivotal points that allow connections between the research area. The clusters #0, #2, and #3 are the most active clusters with the strongest citation bursts (Table 7) [14,4,48-50,52-55]. This implies that these clusters denote where the supreme effort of research in the SMART assay.
In the current study, a comprehensive analysis of the emerging trends in the field of SMART assays from 1984 to 2020 was performed. The analysis reveals an increase in the number of publications in this period, with most of these having been published in the last 10 years, confirming the growing interest in SMART assays. Of note, recently published studies have received fewer citations compared to ancient studies, as it requires time for a study to make an impact. The first observed bloom of TLCS and TGCS has been associated with the first paper introducing the SMART assays in 1984. Thus, the highly cited article is the earliest publication. The second bloom in 1992 has been generated from two influential articles written by Graf and van Schaik (1992) [41] and Frei et al. (1992) [42]. The most prolific author in the SMARTs field is Graf U (Switzerland), with 953 TLCS and 1,716 TGCS. Furthermore, four of his research papers are in the top 10 highly cited articles, and five of them in the top 25 strongest citation bursts showing the interest of researchers in his field. The collaboration between authors could be due to the emergence of interest among researchers. While only one of the prominent clusters is interconnected, it is expected in the future to improve overall collaborative work. The large number of multi-authored documents could be related to various collaborations between countries to expedite the usage of SMART assays. In fact, 35 countries have published about SMART assays. The two influential authors de Andrade HHR and Lehmann M are both from Brazil. Thus, this country had the highest number of publications. The rise in publications from Brazil can also be attributed to the high frequency of collaboration with institutions in Switzerland. Both authors de Andrade HHR and Lehman M were collaborators with Graf U in 2000 to study the genotoxic potential of tannic acid [56]. However, the usage of SMART assays in genotoxicity studies is still ignored in many countries. The most influential journals accounted for 61.5% of all the publications, and this finding illustrated that the distribution of publication was narrow. These influential journals are in Q1(3), Q2(2), Q3(3), and Q4(2) category. To note, five highly cited articles were published in Mutation Research (Q1), two in Mutagenesis (Q2), and one in Environmental and Molecular Mutagenesis (Q2). The first most-cited article by Graf et al. was published in 1984 [11]. This article was published in Environmental Mutagenesis (Q2), ranked at 55th position with only this publication. Therein, the protocol of SMART assay was presented, and several chemicals such as β-propiolactone, 1,2-dibromoethane, aflatoxin B1, diethylnitrosamine, dimethylnitrosamine, mitomycin C, and procarbazine have been identified as mutagens. The second most-cited article by Frei and Wurgler [40] was published in Mutation Research-Environmental Mutagenesis and Related Subjects. This journal ceased publication, to be incorporated in 1997 into Mutation Research-Genetic Toxicology and Environmental Mutagenesis journal (Q3) [57]. In this study, to reduce the risk of inconclusive results, a new statistical test was proposed. The third most-cited article by Graf and van Schaik [41] proposed new strains to improve the visual score. This article was published in Mutation Research (Q1 in 1999) currently known as Mutation Research- Fundamental and Molecular Mechanisms of Mutagenesis (Q3) [57]. The most used keywords allow distinguishing the articles relevant to SMART assays. The three-fields plot showed that Drosophila melanogaster, genotoxicity, SMART, and antigenotoxicity were used by authors from Brazil, Turkey, and Spain. These keywords appear to be generic; yet it was used frequently. Based on thematic analysis, five large clusters were observed. The two clusters ‘somatic mutation’ and ‘nitrogen mustard’ are the two largest and oldest clusters. The cluster ‘copper oxide nanoparticle’ is the youngest cluster. The value of the mean silhouette (S) and the modularity (Q) are 0.9501 and 0.3944, respectively, suggesting reliable and robust results. In the largest cluster labeled ‘somatic mutation’ (#0) with 103 members and which is also the most active cluster, the most actively citing article (a research frontier article) identified polycyclic aromatic hydrocarbons and their derivatives as genotoxic [58]. The most actively cited articles (intellectual-based papers) are Graf et al. (1989) [14], Lindsley and Zimm (1992) [48], and Graf and van Schaik (1992) [41]. Graf et al. [14] is the second top-ranked paper with the strongest citation burst (11.22) and therein the efficiency of the SMART assay was proven by applying it on 30 compounds to evaluate their genotoxic potential. Lindsley and Zimm [48] described and identified the genome of Drosophila. Graf and van Schaik [41] improved new strain to assure better cross. In brief, cluster #0 mainly concentrated on enhancing the SMART protocol to be more practical and accurate. In the second cluster labeled ‘nitrogen mustard’ (#1), the most actively citing paper focused on the effect of tannic acid on nitrogen mustard, mitomycin C, and methylmehanesulfonate [56], whereas the most actively cited paper are Frei and Wurgler [40] and Graf [59]. These two papers focused on the Drosophila model by identifying the sample size required as well as the appropriate age of larvae, depending on the mutagens to avoid inconclusive results. It can be concluded that cluster #1 focused on the incorporation of a new control positive (nitrogen mustard) into SMART assay. Not surprisingly that the two largest clusters cover the most interests as both focused on the improvement of SMART assays protocol. In the third cluster labeled ‘vivo model’ (#2) the most actively citing paper was written by Carmona et al. [60] to evaluate the genotoxic potential of titanium dioxide anatase nanoparticles. Demir et al. [49] and Vales et al. [52] are the most-cited paper. Demir et al. [49] analyzed the genotoxic potential of silver nanoparticles whereas Vales et al. [52] evaluated the genotoxic effect of cobalt nanoparticles. It seems that in cluster #2 nanoparticles have gradually attracted the attention of scholars. Although the most-cited and citing papers were mainly dedicated to the genotoxic effect of nanoparticles, most of members treated various compounds, justifying thereby the label ‘vivo model’ instead of nanoparticles. In the fourth cluster labeled ‘doxorubicin-induced somatic mutation’ (#3), all the intellectual bases are related to cancer. Fragiorge et al. [53] analyzed the antigenotoxicity effect of ascorbic acid on doxorubicin. Doxorubicin is an antibiotic to treat human cancers which generally induces genotoxicity by oxidative damage [61]. Bishop and Schiestl [54] studied the function of homologous recombination in cancer. Similarly, the citing paper focused on the use of the herbal extract of ginseng to inhibit the genotoxic effect of doxorubicin [62]. In brief, cluster #3 was concentrated on resolving the doxorubicin-induced genotoxicity. The fifth cluster labeled ‘copper oxide nanoparticle’ (#4) is considered a persistent cluster denoting the continuity of an existing trend. In this cluster, the research frontier assessed the effect of copper oxide nanoparticles [63]. The metal oxide nanoparticles possess a redox property suggesting an antigenotoxic and anticarcinogenic potential [21]. The intellectual base emphasis on nanomaterials. Alaraby et al. [64] discussed the side effect of nanomaterials. Carmona et al. [60] presented the titanium dioxide anatase nanoparticles, being generally used in pharmaceuticals and cosmetics. This nanomaterial can generate oxidative stress, and subsequently genotoxicity [65]. Interestingly, this paper was a research frontier in cluster #2. It is worth mentioning, the first top-ranked burst item Frei and Wurgler (16.12) [47] is in a small and old cluster (1986) labeled ‘antiparasitic nitrofuran’ (#6) with 45 members. In sum, the research frontiers share sometimes the same theme with the intellectual bases since most often the continuation and the growth of intellectual base are the research fronts. The present study has certain limitations. Data were extracted from the WOS database; other databases were not considered. The database WOS is always updating, even with recently published papers, top publications are high enough that including recent paper would not have an influence on this research. The proceedings papers, book chapters, and early access are not included. Even though book chapters were excluded, the book by Wurgler and Vogel (1986) [12] describing the eye-spot assay was not indexed in the WOS database. The researchers tried carefully to include a maximum of relevant keywords; however, two studies Martinez-Valdivieso et al. (2017) [66] and Fernandez-Bedmar and Alonso-Moraga (2016) [67] have been missed. The absence of these papers is due to keyword search where none of the relevant used keywords were present in the title, neither in the abstract and/or the keywords section. The number of authors may differ since some authors published articles with a different initial of the first name. This led to some authors being separated into two authors. Even with these limitations, the current study on SMART assays can provide a directive to researchers to find influential articles, journals, and authors to assure collaboration and to reduce the research gaps.
In this study, a comprehensive bibliometric analysis was performed on SMART assays literature. Most of the publications were published in the last 10 years, mainly from Brazil, especially from the Federal University of Uberlândia. Notably, multiple authors were publishing papers on this field and mostly in ‘Food and Chemical Toxicology’ and ‘Mutation Research Genetic Toxicology and Environmental Mutagenesis’ journals. By analyzing the emerging trends, the nanoparticle area represents a persistent cluster. This area is expected to draw more attention. Finally, furthers studies should assess the records present on other databases to find out if the same trends for SMART assays are present.
Notes
Fig. 2.
Evolution of the number of articles and citations over the years. Recs, number of publications; TGCS, total global citation score; TLCS, total local citation score.
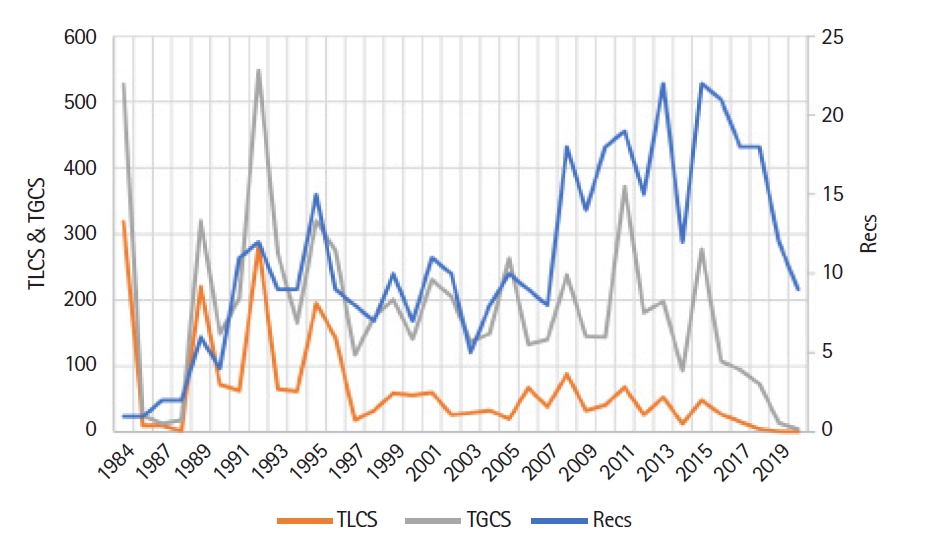
Fig. 3.
Co-authorship network (Walktrap algorithm, association normalization, 20 nodes, 1 minimum edge, created by Biblioshiny).
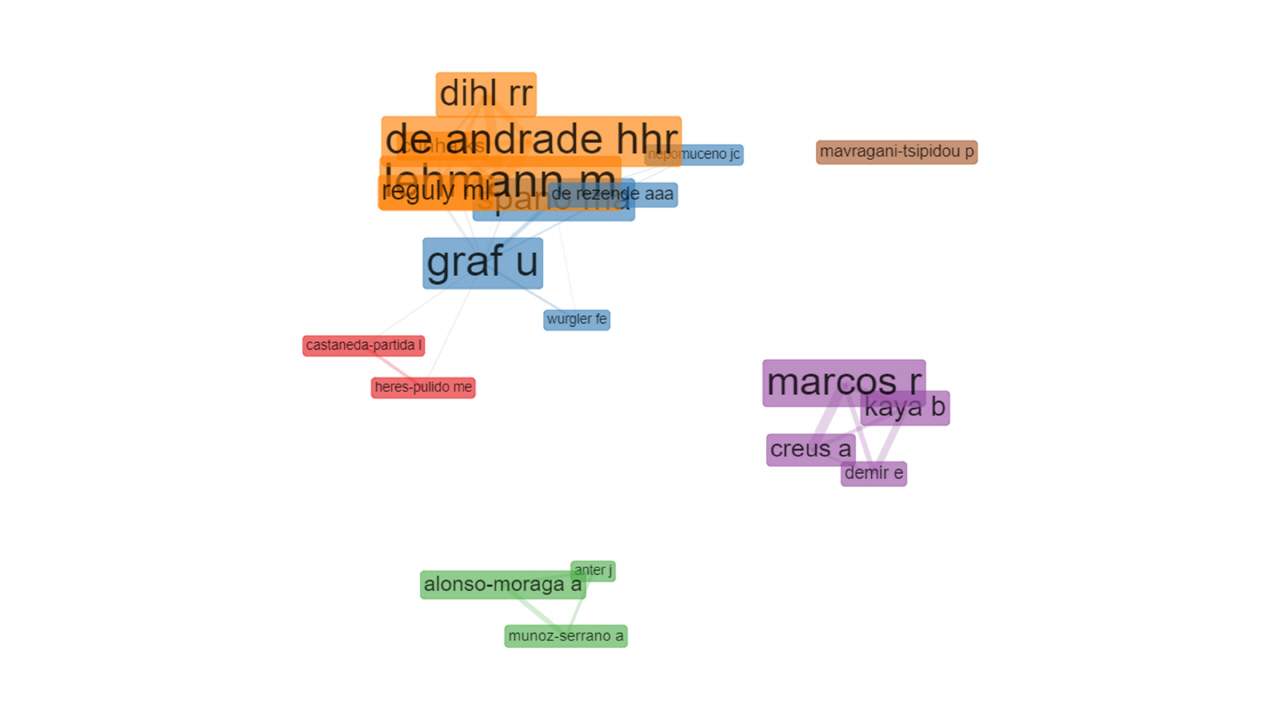
Fig. 5.
Co-occurrence network of author keywords. The green and the red clusters are the strongest ones (12 words) followed by the blue cluster (10 words). The top word is Drosophila melanogaster with the maximum occurrence in somatic mutation and recombination test assays literature.
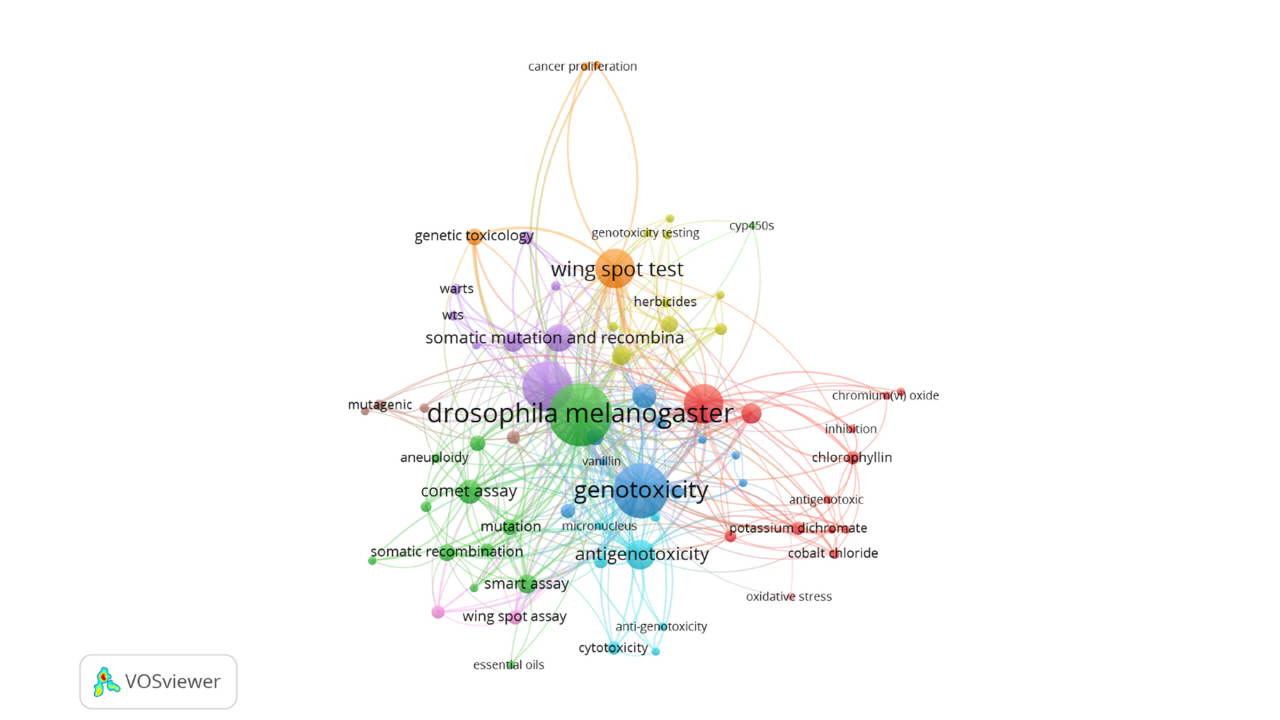
Fig. 7.
Co-citation network of SMART assays field. Each link colors indicate a given time slice. The oldest co-citation relationships are visualized as dark blue, whereas yellow links presented articles that are recently co-cited (created by CiteSpace). SMART, somatic mutation and recombination test.
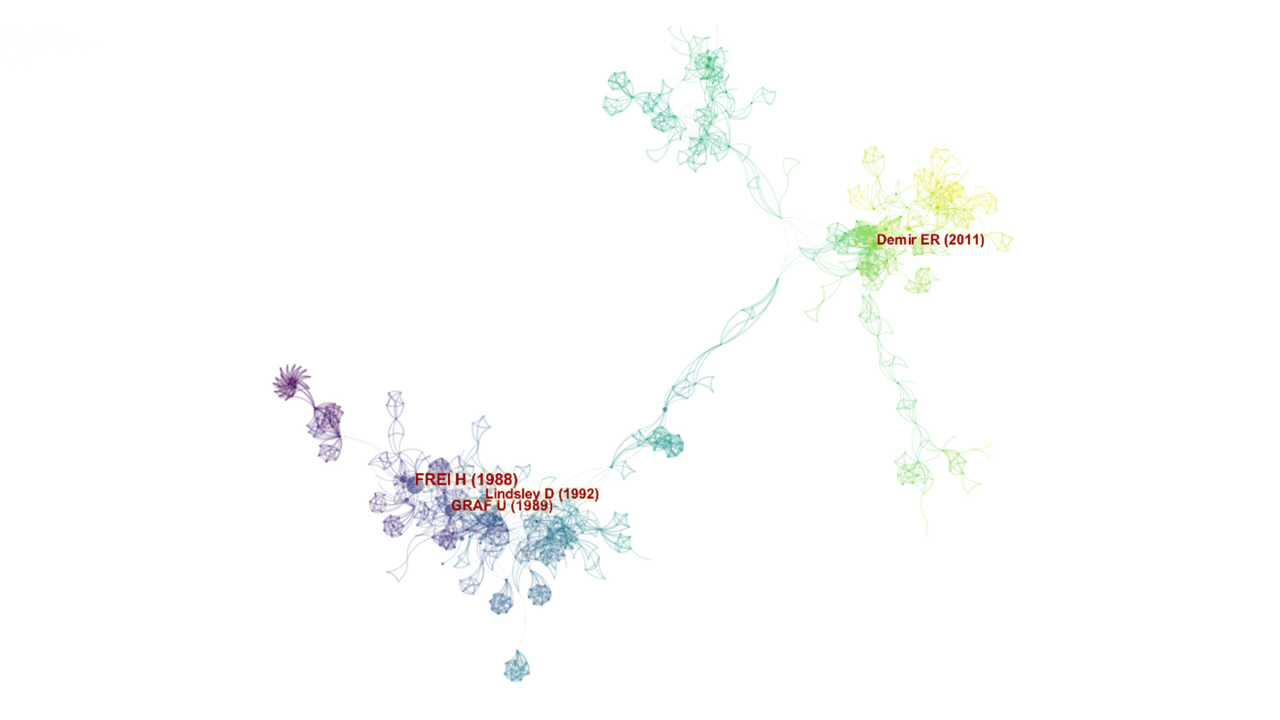
Fig. 8.
The top 25 references of somatic mutation and recombination test assays literature. The blue lines represent the time span (1984‒2020), the red lines denote the period of the bursts, strength indicates the burst strength (created by CiteSpace).

Fig. 9.
Timeline view of the largest five clusters of document co-citation based on one-year time slices from 1984 to 2020. The order of cluster from top to down is related to their size, and the arrangement of clusters from left to right denotes their time distribution. The frequency of co-citations is proportional to the size of nodes. The red rings around the nodes represent the citation bursts (created by CiteSpace).

Table 1.
The top 10 impactful authors ranked by Recs
Table 2.
The most productive institutions and countries ranked by Recs
Table 3.
The most productive journals ranked by Recs
| Journal | Recs | TLCS | TLCS/t | TGCS | TGCS/t | IF (2020) |
|---|---|---|---|---|---|---|
| Food and Chemical Toxicology | 53 | 165 | 15.8 | 714 | 69.78 | 6.025 (Q1) |
| Mutation Research-Genetic Toxicology and Environmental Mutagenesis | 53 | 250 | 17.55 | 1,085 | 81.73 | 2.873 (Q3) |
| Environmental and Molecular Mutagenesis | 31 | 213 | 11.24 | 511 | 29.03 | 3.216 (Q2) |
| Mutation Research | 30 | 610 | 20.28 | 1,128 | 37.78 | 2.11 (Q1)a |
| Mutagenesis | 20 | 187 | 7.99 | 440 | 18.44 | 3.000 (Q2) |
| Fresenius Environmental Bulletin | 17 | 34 | 3.35 | 53 | 5.78 | 0.489 (Q4) |
| Chemosphere | 10 | 30 | 3.12 | 177 | 19.19 | 7.086 (Q1) |
| Genetics and Molecular Biology | 9 | 28 | 2.2 | 63 | 6.01 | 1.771 (Q3) |
| Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesisb | 9 | 28 | 1.12 | 114 | 5.78 | 2.433 (Q3) |
| Toxicology and Industrial Health | 9 | 17 | 2.85 | 68 | 11.73 | 2.273 (Q4) |
Table 4.
The top 10 highly cited articles ranked by LCS
| Title | Author | Source | Year | LCS | GCS |
|---|---|---|---|---|---|
| Somatic mutation and recombination test in Drosophila melanogaster | Graf et al. [11] | Environmental Mutagenesis | 1984 | 319 | 527 |
| Optimal experimental-design and sample-size for the statistical evaluation of data from somatic mutation and recombination tests (SMART) in Drosophila | Frei and Wurgler [40] | Mutation Research-Environmental Mutagenesis and Related Subjects | 1995 | 133 | 156 |
| Improved high bioactivation cross for the wing somatic mutation and recombination test in Drosophila melanogaster | Graf and van Schaik [41] | Mutation Research | 1992 | 132 | 159 |
| 30 Compounds tested in the Drosophila wing spot-test | Graf et al. [14] | Mutation Research | 1989 | 121 | 165 |
| The genotoxicity of the anticancer drug mitoxantrone in somatic and germ-cells of Drosophila melanogaster | Frei et al. [42] | Mutation Research | 1992 | 85 | 100 |
| Induction of somatic mutation and recombination by four inhibitors of eukaryotic topoisomerases assayed in the wing spot test of Drosophila melanogaster | Frei and Wurgler [43] | Mutagenesis | 1996 | 42 | 56 |
| Metabolism of promutagens catalyzed by Drosophila melanogaster Cyp6a2 enzyme in Saccharomyces cerevisiae | Saner et al. [44] | Environmental and Molecular Mutagenesis | 1996 | 41 | 91 |
| Genotoxicity testing of antiparasitic nitrofurans in the Drosophila wing somatic mutation and recombination test | Moraga and Graf [45] | Mutagenesis | 1989 | 37 | 47 |
| Drosophila wing-spot test - improved detectability of genotoxicity of polycyclic aromatic-hydrocarbons | Frolich and Wurgler [46] | Mutation Research | 1990 | 29 | 50 |
Table 5.
The top five critical articles in SMART assays
| Cited frequency | Title | Author | Year | Betweenness centrality |
|---|---|---|---|---|
| 30 | Statistical methods to decide whether mutagenicity test data from Drosophila assays indicate a positive, negative, or inconclusive result | Frei and Wurgler [47] | 1988 | 0.08 |
| 21 | 30 Compounds tested in the Drosophila wing spot-test | Graf et al. [14] | 1989 | 0.13 |
| 19 | The genome of Drosophila melanogaster | Lindsley and Zimm [48] | 1992 | 0.08 |
| 19 | Genotoxic analysis of silver nanoparticles in Drosophila | Demir et al. [49] | 2011 | 0.12 |
| 14 | Improved high bioactivation cross for the wing somatic mutation and recombination test in Drosophila melanogaster | Graf and van Schaik [41] | 1992 | 0.12 |
Table 6.
The top five clusters in the literature
Table 7.
The top three articles in clusters #0, #2, and #3 with the strongest citation bursts
| ID | Burst | Title | Author | Year |
|---|---|---|---|---|
| 0 | 11.22 | 30 Compounds tested in the Drosophila wing spot-test | Graf et al. [14] | 1989 |
| 10.07 | The genome of Drosophila melanogaster | Lindsley and Zimm [48] | 1992 | |
| 7.39 | Improved high bioactivation cross for the wing somatic mutation and recombination test in Drosophila melanogaster | Graf and van Schaik [41] | 1992 | |
| 2 | 9.31 | Genotoxic analysis of silver nanoparticles in Drosophila | Demir et al. [49] | 2011 |
| 7.21 | Genotoxicity of cobalt nanoparticles and ions in Drosophila | Vales el al. [52] | 2013 | |
| 5.47 | Proposal of an in vivo comet assay using haemocytes of Drosophila melanogaster | Carmona et al. [50] | 2011 | |
| 3 | 5.91 | Modulatory effects of the antioxidant ascorbic acid on the direct genotoxicity of doxorubicin in somatic cells of Drosophila melanogaster | Fragiorge et al. [53] | 2007 |
| 5.80 | Role of homologous recombination in carcinogenesis | Bishop and Schiestl [54] | 2003 | |
| 5.73 | Protective effects of a mixture of antioxidant vitamins and minerals on the genotoxicity of doxorubicin in somatic cells of Drosophila melanogaster | Costa and Nepomuceno [55] | 2006 |
References
1. Bilal M, Rasheed T, Nabeel F, Iqbal HM, Zhao Y. Hazardous contaminants in the environment and their laccase-assisted degradation: a review. J Environ Manage 2019;234:253–264.


2. Kopp B, Zalko D, Audebert M. Genotoxicity of 11 heavy metals detected as food contaminants in two human cell lines. Environ Mol Mutagen 2018;59:202–210.


3. Aiassa D. Genotoxic risk in human populations exposed to pesticides. In: Genotoxicity: A Predictable Risk to Our Actual World (Larramendy ML, Soloneski S, eds.). London: InTech, 2018. pp. 95–112.
4. Khan S, Anas M, Malik A. Mutagenicity and genotoxicity evaluation of textile industry wastewater using bacterial and plant bioassays. Toxicol Rep 2019;6:193–201.



5. Hayes AW, Sahu SC. Genotoxicity of engineered nanomaterials found in the human environment. Curr Opin Toxicol 2020;19:68–71.

6. OECD Guidelines for the testing of chemicals. Test No. 487: In vitro mammalian cell micronucleus test. Paris: Organisation for Economic Co-operation and Development, 2016.
7. Barnes JL, Zubair M, John K, Poirier MC, Martin FL. Carcinogens and DNA damage. Biochem Soc Trans 2018;46:1213–1224.



8. Tiwari V, Wilson DM 3rd. DNA damage and associated DNA repair defects in disease and premature aging. Am J Hum Genet 2019;105:237–257.



9. Corvi R, Madia F. In vitro genotoxicity testing: can the performance be enhanced? Food Chem Toxicol 2017;106:600–608.


10. Drosopoulou E, Vlastos D, Efthimiou I, Kyrizaki P, Tsamadou S, Anagnostopoulou M, et al. In vitro and in vivo evaluation of the genotoxic and antigenotoxic potential of the major Chios mastic water constituents. Sci Rep 2018;8:12200.




11. Graf U, Wurgler FE, Katz AJ, Frei H, Juon H, Hall CB, et al. Somatic mutation and recombination test in Drosophila melanogaster. Environ Mutagen 1984;6:153–188.


12. Wurgler FE, Vogel EW. In vivo mutagenicity testing using somatic cells of Drosophila melanogaster. In: Chemical Mutagens, Principles and Methods for Their Detection. Vol. 10 (Serres FJ, ed.). New York: Plenum Press, 1986. pp. 1–72.
13. Spano MA, Frei H, Wurgler FE, Graf U. Recombinagenic activity of four compounds in the standard and high bioactivation crosses of Drosophila melanogaster in the wing spot test. Mutagenesis 2001;16:385–394.


14. Graf U, Frei H, Kagi A, Katz AJ, Wurgler FE. Thirty compounds tested in the Drosophila wing spot test. Mutat Res 1989;222:359–373.


15. Frolich A, Wurgler FE. New tester strains with improved bioactivation capacity for the Drosophila wing-spot test. Mutat Res 1989;216:179–187.


16. Vogel EW, Nivard MJ, Zijlstra JA. Variation of spontaneous and induced mitotic recombination in different Drosophila populations: a pilot study on the effects of polyaromatic hydrocarbons in six newly constructed tester strains. Mutat Res 1991;250:291–298.


17. Gaivao I, Ferreira J, Maria Sierra L. The w/w + somatic mutation and recombination test (SMART) of Drosophila melanogaster for detecting antigenotoxic activity. In: Genotoxicity and Mutagenicity: Mechanisms and Test Methods (Soloneski S, Larramendy ML, eds.). London: IntechOpen, 2021. pp. 111–145.
18. Kaya B, Marcos R, Yanikoglu A, Creus A. Evaluation of the genotoxicity of four herbicides in the wing spot test of Drosophila melanogaster using two different strains. Mutat Res 2004;557:53–62.


19. de Morais CR, Carvalho SM, Carvalho Naves MP, Araujo G, de Rezende AA, Bonetti AM, et al. Mutagenic, recombinogenic and carcinogenic potential of thiamethoxam insecticide and formulated product in somatic cells of Drosophila melanogaster. Chemosphere 2017;187:163–172.


20. Castaneda-Sortibran AN, Flores-Loyola C, Martinez-Martinez V, Ramirez-Corchado MF, Rodriguez-Arnaiz R. Herbicide genotoxicity revealed with the somatic wing spot assay of Drosophila melanogaster. Rev Int Contam Ambient 2019;35:295–305.

21. Alaraby M, Hernandez A, Marcos R. Copper oxide nanoparticles and copper sulphate act as antigenotoxic agents in Drosophila melanogaster. Environ Mol Mutagen 2017;58:46–55.


22. Avalos A, Haza AI, Mateo D, Morales P. In vitro and in vivo genotoxicity assessment of gold nanoparticles of different sizes by comet and SMART assays. Food Chem Toxicol 2018;120:81–88.


23. Fernandez-Bedmar Z, Anter J, Alonso Moraga A. Anti/genotoxic, longevity inductive, cytotoxic, and clastogenic-related bioactivities of tomato and lycopene. Environ Mol Mutagen 2018;59:427–437.


24. Sukprasansap M, Sridonpai P, Phiboonchaiyanan PP. Eggplant fruits protect against DNA damage and mutations. Mutat Res 2019;813:39–45.


25. Ertugrul H, Yalcin B, Gunes M, Kaya B. Ameliorative effects of melatonin against nano and ionic cobalt induced genotoxicity in two in vivo Drosophila assays. Drug Chem Toxicol 2020;43:279–286.


26. Ferreira J, Marques A, Abreu H, Pereira R, Rego A, Pacheco M, et al. Red seaweeds Porphyra umbilicalis and Grateloupia turuturu display antigenotoxic and longevity-promoting potential in Drosophila melanogaster. Eur J Phycol 2019;54:519–530.

27. de Moraes Filho AV, de Jesus Silva Carvalho C, Vercosa CJ, Goncalves MW, Rohde C, de Melo ES, et al. In vivo genotoxicity evaluation of efavirenz (EFV) and tenofovir disoproxil fumarate (TDF) alone and in their clinical combinations in Drosophila melanogaster. Mutat Res Genet Toxicol Environ Mutagen 2017;820:31–38.


28. Naves MP, de Morais CR, Spano MA, de Rezende AA. Mutagenicity and recombinogenicity evaluation of bupropion hydrochloride and trazodone hydrochloride in somatic cells of Drosophila melanogaster. Food Chem Toxicol 2019;131:110557.


29. Chen-Chen L, de Jesus Silva Carvalho C, de Moraes Filho AV, Veras JH, Cardoso CG, Bailao E, et al. Toxicity and genotoxicity induced by abacavir antiretroviral medication alone or in combination with zidovudine and/or lamivudine in Drosophila melanogaster. Hum Exp Toxicol 2019;38:446–454.


30. Ellegaard O, Wallin JA. The bibliometric analysis of scholarly production: How great is the impact? Scientometrics 2015;105:1809–1831.



31. van Nunen K, Li J, Reniers G, Ponnet K. Bibliometric analysis of safety culture research. Saf Sci 2018;108:248–258.

32. Liu W, Tang L, Gu M, Hu G. Feature report on China: a bibliometric analysis of China-related articles. Scientometrics 2015;102:503–517.

33. Pons P, Latapy M. In: Computer and Information Sciences - ISCIS 2005. Lecture Notes in Computer Science, Vol. 3733 (Yolum P, Gungor T, Gurgen F, Ozturan C, eds.). Berlin: Springer, 2005. pp. 284-293.
34. Garfield E, Paris SW, Stock WG. HistCite: a software tool for informetric analysis of citation linkage. Information-Wissenschaft und Praxis 2006;57:391–400.
35. Aria M, Cuccurullo C. bibliometrix: an R-tool for comprehensive science mapping analysis. J Informetr 2017;11:959–975.

36. Liu Y, Sun T, Yang L. Evaluating the performance and intellectual structure of construction and demolition waste research during 2000-2016. Environ Sci Pollut Res Int 2017;24:19259–19266.


37. Wei F, Grubesic TH, Bishop BW. Exploring the GIS knowledge domain using CiteSpace. The Prof Geogr 2015;67:374–384.

38. Chen C. CiteSpace II: detecting and visualizing emerging trends and transient patterns in scientific literature. J Am Soc Inf Sci Technol 2006;57:359–377.

39. Chen C, Chen Y, Horowitz M, Hou H, Liu Z, Pellegrino D. Towards an explanatory and computational theory of scientific discovery. J Informetr 2009;3:191–209.

40. Frei H, Wurgler FE. Optimal experimental design and sample size for the statistical evaluation of data from somatic mutation and recombination tests (SMART) in Drosophila. Mutat Res 1995;334:247–258.


41. Graf U, van Schaik N. Improved high bioactivation cross for the wing somatic mutation and recombination test in Drosophila melanogaster. Mutat Res 1992;271:59–67.


42. Frei H, Clements J, Howe D, Wurgler FE. The genotoxicity of the anti-cancer drug mitoxantrone in somatic and germ cells of Drosophila melanogaster. Mutat Res 1992;279:21–33.


43. Frei H, Wurgler FE. Induction of somatic mutation and recombination by four inhibitors of eukaryotic topoisomerases assayed in the wing spot test of Drosophila melanogaster. Mutagenesis 1996;11:315–325.


44. Saner C, Weibel B, Wurgler FE, Sengstag C. Metabolism of promutagens catalyzed by Drosophila melanogaster CYP6A2 enzyme in Saccharomyces cerevisiae. Environ Mol Mutagen 1996;27:46–58.


45. Moraga AA, Graf U. Genotoxicity testing of antiparasitic nitrofurans in the Drosophila wing somatic mutation and recombination test. Mutagenesis 1989;4:105–110.


46. Frolich A, Wurgler FE. Drosophila wing-spot test: improved detectability of genotoxicity of polycyclic aromatic hydrocarbons. Mutat Res 1990;234:71–80.


47. Frei H, Wurgler FE. Statistical methods to decide whether mutagenicity test data from Drosophila assays indicate a positive, negative, or inconclusive result. Mutat Res 1988;203:297–308.


48. Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. San Diego: Academic Press, 1992.
49. Demir E, Vales G, Kaya B, Creus A, Marcos R. Genotoxic analysis of silver nanoparticles in Drosophila. Nanotoxicology 2011;5:417–424.


50. Carmona ER, Guecheva TN, Creus A, Marcos R. Proposal of an in vivo comet assay using haemocytes of Drosophila melanogaster. Environ Mol Mutagen 2011;52:165–169.


51. Chen C, Dubin R, Kim MC. Emerging trends and new developments in regenerative medicine: a scientometric update (2000 - 2014). Expert Opin Biol Ther 2014;14:1295–1317.


52. Vales G, Demir E, Kaya B, Creus A, Marcos R. Genotoxicity of cobalt nanoparticles and ions in Drosophila. Nanotoxicology 2013;7:462–468.


53. Fragiorge EJ, Spano MA, Antunes LM. Modulatory effects of the antioxidant ascorbic acid on the direct genotoxicity of doxorubicin in somatic cells of Drosophila melanogaster. Genet Mol Biol 2007;30:449–455.

54. Bishop AJ, Schiestl RH. Role of homologous recombination in carcinogenesis. Exp Mol Pathol 2003;74:94–105.


55. Costa WF, Nepomuceno JC. Protective effects of a mixture of antioxidant vitamins and minerals on the genotoxicity of doxorubicin in somatic cells of Drosophila melanogaster. Environ Mol Mutagen 2006;47:18–24.


56. Lehmann M, Graf U, Reguly ML, Rodrigues De Andrade HH. Interference of tannic acid on the genotoxicity of mitomycin C, methylmethanesulfonate, and nitrogen mustard in somatic cells of Drosophila melanogaster. Environ Mol Mutagen 2000;36:195–200.


57. Timeline of Mutation Research journals since 1964. Amsterdam: Elsevier, 2019. Accessed 2021 Nov 2. Available from: https://www.journals.elsevier.com/dna-repair/history-of-mutation-research/timeline-of-mutation-research-journals-since-1964.
58. Delgado-Rodriguez A, Ortiz-Marttelo R, Graf U, Villalobos-Pietrini R, Gomez-Arroyo S. Genotoxic activity of environmentally important polycyclic aromatic hydrocarbons and their nitro derivatives in the wing spot test of Drosophila melanogaster. Mutat Res 1995;341:235–247.


59. Graf U. Analysis of the relationship between age of larvae at mutagen treatment and frequency and size of spots in the wing somatic mutation and recombination test in Drosophila melanogaster. Experientia 1995;51:168–173.


60. Carmona ER, Escobar B, Vales G, Marcos R. Genotoxic testing of titanium dioxide anatase nanoparticles using the wing-spot test and the comet assay in Drosophila. Mutat Res Genet Toxicol Environ Mutagen 2015;778:12–21.


61. Hajra S, Patra AR, Basu A, Bhattacharya S. Prevention of doxorubicin (DOX)-induced genotoxicity and cardiotoxicity: effect of plant derived small molecule indole-3-carbinol (I3C) on oxidative stress and inflammation. Biomed Pharmacother 2018;101:228–243.


62. Pereira DG, Antunes LM, Graf U, Spano MA. Protection by Panax ginseng C.A. Meyer against the genotoxicity of doxorubicin in somatic cells of Drosophila melanogaster. Genet Mol Biol 2008;31:947–955.

63. Carmona ER, Inostroza-Blancheteau C, Obando V, Rubio L, Marcos R. Genotoxicity of copper oxide nanoparticles in Drosophila melanogaster. Mutat Res Genet Toxicol Environ Mutagen 2015;791:1–11.


64. Alaraby M, Annangi B, Marcos R, Hernandez A. Drosophila melanogaster as a suitable in vivo model to determine potential side effects of nanomaterials: a review. J Toxicol Environ Health B Crit Rev 2016;19:65–104.


65. Liao F, Chen L, Liu Y, Zhao D, Peng W, Wang W, et al. The size-dependent genotoxic potentials of titanium dioxide nanoparticles to endothelial cells. Environ Toxicol 2019;34:1199–1207.


- TOOLS
-
METRICS

-
- 6 Crossref
- 0 Scopus
- 4,870 View
- 150 Download
- Related articles in GNI