Coronaviruses (CoVs) generally disturb human beings' respiratory tract and other mammals that causes severe respiratory infections. A previous study reveals that two extremely pathogenic human CoVs, including severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), growing from viral family, have led to worldwide epidemics at different times [
1,
2]. According to the World Health Organization, as of 15 Jan 2021, CoV had 13.1 million diagnosed cases causing 1.98 million deaths throughout the world [
3]. A comparative study indicates that the ORF8 protein of SARS-CoV attained from host genes during evolution [
4]. Comprehensive genomic analysis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strains and its closely related coronavirus strains shows that the ratio of nucleotide to amino acid substitutions of the spike gene is higher [
5]. Lower R0 values of MERS-CoV full genome assist in tracking transmission and multiple mutations [
6]. Similarly, the complete genomic sequence of the porcine hamagglutinating encephalalomylitis virus discloses the existence of a truncated group ns2 gene [
7]. Turkey coronavirus that has divergence within spike protein and provided the evidence of recombination that can directly lead to new coronaviruses [
8]. The analysis was performed on 93 complete genomes of SARS-CoV-2 from the GISAID (Global Initiative for Sharing All Influenza Data) database to explore the evolution and human-to-human transmissions of SARS-CoV-2 [
9]. The recombination study proved that the newly discovered MERS-CoV had obtained their spike genes from a bat coronavirus, which provide significant proof that bats represent MERS-CoV evolutionary origins [
10]. Simulated 219 camels and human MERS-CoV genome sequences available in GenBank for Phylogenetic analysis showed that clade B divided into B1 to B6 (each containing both human and camel strains) [
11]. Viral spike glycoprotein, which recognizes a cell surface receptor, supports that the SARS-CoV-2 a recombinant of the bat coronavirus and an unknown origin coronavirus. Higher sequence homology found between SARS-CoV-2 to bat CoV RaTG13 suggested that the Chinese chrysanthemum bat is the origin of SARS-CoV2 in China [
12,
13]. The Vietnam B-CoV, Cuban, and Chinese strains have high nucleotide sequence similarity due to same cluster [
14]. Experimental data show that SARS-CoV-2 originate from bat CoV RaTG13 under the positive selection hypothesis model having common ancestor [
15]. The inequality between nonsynonymous to synonymous changes simulates using capsid protein of human herpes virus to understand the evolution [
16]. MEGA software implements numerous statistical techniques for nucleotide substitution models and estimates evolutionary rates [
17]. The nonsynonymous mutations of SARS-CoV-2 were isolated and evaluated for surface glycoprotein spike for amino acid alterations [
18]. The protein sequence similarity of pangolin-hCoV and bat-hCoV with human coronavirus was higher than their nucleotide similarity, denoting the occurrence of more synonymous mutations in the genome [
19]. It has been observed that, due to the structure of the genetic code, nonsynonymous transitions are recessive than transversions to leads radical changes in amino acids [
20]. In this simulative study, four nucleotide substitution methods simulated for revealing dN/dS and improved method, assumes that the substitution rate is not equal.
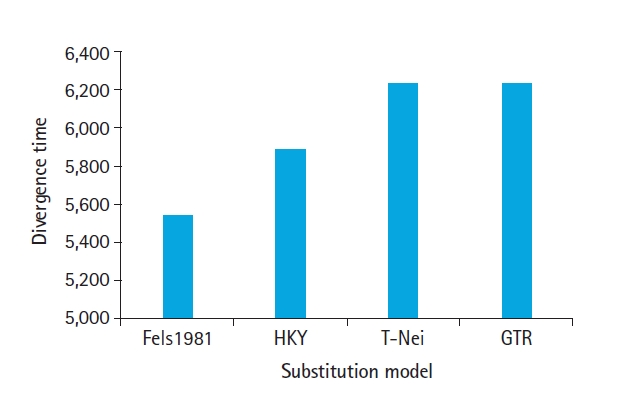
The purpose of the present work is to estimate the synonymous and nonsynonymous substitution in the genome of MERS-CoV, SARS-CoV, and SARS-CoV-2 with an objective to (1) normalization dN-dS value using codon through HyPhy using maximum likelihood approach (2) Z-test of selection for estimation of positive or neutral evolution using different transition/transversion ratio (tr/tv) (3) estimate the effect of different substitution models on divergence time.