|
 |
- Search
| Genomics Inform > Volume 18(2); 2020 > Article |
|
Abstract
Named entity recognition tools are used to identify mentions of biomedical entities in free text and are essential components of high-quality information retrieval and extraction systems. Without good entity recognition, methods will mislabel searched text and will miss important information or identify spurious text that will frustrate users. Most tools do not capture non-contiguous entities which are separate spans of text that together refer to an entity, e.g., the entity ŌĆ£type 1 diabetesŌĆØ in the phrase ŌĆ£type 1 and type 2 diabetes.ŌĆØ This type is commonly found in biomedical texts, especially in lists, where multiple biomedical entities are named in shortened form to avoid repeating words. Most text annotation systems, that enable users to view and edit entity annotations, do not support non-contiguous entities. Therefore, experts cannot even visualize non-contiguous entities, let alone annotate them to build valuable datasets for machine learning methods. To combat this problem and as part of the BLAH6 hackathon, we extended the TextAE platform to allow visualization and annotation of non-contiguous entities. This enables users to add new subspans to existing entities by selecting additional text. We integrate this new functionality with TextAEŌĆÖs existing editing functionality to allow easy changes to entity annotation and editing of relation annotations involving non-contiguous entities, with importing and exporting to the PubAnnotation format. Finally, we roughly quantify the problem across the entire accessible biomedical literature to highlight that there are a substantial number of non-contiguous entities that appear in lists that would be missed by most text mining systems.
Information extraction and retrieval methods are essential tools to enable scientists to find and read the appropriate papers to enable discoveries. Many of these methods require identifying mentions of specific biomedical entities in the text and make use of named entity recognition (NER) tools for this task. Most entities are represented by a single span of text, e.g., the name of a drug. However, some entities are represented by multiple spans of text that are separated by other words and together identify the entity, for example, the separate words ŌĆ£skinŌĆØ and ŌĆ£cancerŌĆØ in ŌĆ£skin and lung cancer.ŌĆØ These are known as non-contiguous, or discontiguous entities. Table 1 illustrates several more examples from public text mining resources. It should be noted that non-contiguous entities are different from anaphora or coreference resolution, in which multiple spans refer to the same entity separately and do not work together to identify the entity.
Robust annotation tools that are capable of annotating non-contiguous entities are important so that valuable entity information is not missed. These tools are needed to create corpora with non-contiguous entities that can be used as training data for machine learning-based NER methods and also evaluate all NER methods fairly. The leading NER methods frequently use machine-learning methods such as conditional random fields (CRF) that are incapable of capturing non-contiguous entities without additional postprocessing. Popular tools such as BANNER [1], tmChem [2], and DNorm [3] do not support non-contiguous entities.
Many annotation tools have been developed for manual tagging of entities within a document for the biological domain and other domains. A detailed recent review of the strengths and weaknesses of different methods can be found in Neves and SevaŌĆÖs study [4]. To gauge the support for non-contiguous entities, we manually tested the 15 tools selected in that review with an overview shown in Table 2. We were able to run all but one, PDFAnno which displayed an error message that others have reported on Github. We found that only 2 support non-contiguous entities, BRAT [5], and Catma. Furthermore the AlvisAE [6] tool that was not included in the review also supports non-contiguous entity annotation. We suggest that more tools need to provide support for non-contiguous entities.
To that goal, we describe the addition of non-contiguous entity support to TextAE. TextAE is an annotation platform that forms part of the PubAnnotation system for storing and editing text annotations [7]. It is a Node.js web component that accepts text annotations in PubAnnotation JSON format. The PubAnnotation format currently has support for non-contiguous entities but are converted to an alternative representation when edited using the current release of TextAE, known as the chaining representation. This representation converts an entity that contains multiple spans to multiple entities and links them with a relation with type ŌĆ£_lexicallyChainedTo.ŌĆØ This representation is time-consuming to edit and visualizes poorly. Fig. 1 illustrates the current representation of three non-contiguous entities within a sentence using the chaining method. With the current TextAE interface, it is time-consuming to annotate each entity. Assuming TextAE has been set up with appropriate entity and relation types, for each non-contiguous entity, it requires creating two entities (2 mouse clicks), designating one entity with the type ŌĆ£_FRAGMENTŌĆØ (2 clicks), switching to the relation mode (1 click), creating a relation between the two entities (2 clicks), changing its type to ŌĆ£_lexicallyChainedToŌĆØ (2 clicks) and switching back to Term Mode to continue entity annotation (1 click). Even for a TextAE power user, ten clicks for each non-contiguous entity is time-consuming for a large-scale annotation and produces an unwieldy result which is not visually clear.
In this paper, we describe our solution of an extension to the existing TextAE annotation platform to provide seamless support for annotating non-contiguous entities. Finally, we provide evidence of the widespread nature of non-contiguous entities in the biomedical literature using a rule-based extraction system to roughly quantify the scale of non-contiguous entities across all PubMed abstracts and accessible PubMed Central full-text papers.
To develop improved methods to capture non-contiguous entities, well-annotated data needs to be prepared and examined that contain non-contiguous entities. We extend the TextAE annotation platform that is part of the PubAnnotation system [7]. This enables annotation of entities with multiple spans as shown in Fig. 2 with a new subspan mode. The user can select new spans of text that will be added to an existing entity and displayed clearly.
The first task for implementation was changing the underlying span model in TextAE so that all spans are represented as a list of subspans. We dynamically check the input annotation data (in PubAnnotation format) to check if an entity has a single span, or a list of spans, and convert all entities to contain lists of spans, even for single spans. Previously, spans were rendered using a single HTML span tag around the section with appropriate CSS styling to identify the span as an entity. To visualize the new subspans, we removed the styling from the span class, and create subspans for each part of the span and transferred the stylings to the subspans. TextAE implements an Undo/Redo system so changes were required across the codebase to enable the existing functionality to work with the new underlying data structure and allow entities to be manipulated as before.
A new toggleable button (Add subspan) was added to the toolbar. When this button is toggled, any new spans that are selected by the user are added to the previously selected entity. This requires checking that new subspans were compatible with the structure that is enforced by the HTML page. This means that spans and subspans cannot intersect unless one is contained within the other entirely. This means that in the snippet: ŌĆ£breast cancer geneŌĆØ, it would not be possible for ŌĆ£breast cancerŌĆØ and ŌĆ£cancer geneŌĆØ to be tagged as entities. However ŌĆ£breast cancerŌĆØ and ŌĆ£cancerŌĆØ could be tagged as ŌĆ£cancerŌĆØ is fully contained within ŌĆ£breast cancer.ŌĆØ We have not come across use-cases where this functionality is currently needed but cannot discount the potential of this limitation. Fig. 2 illustrates the user interface with an example of non-contiguous entities.
TextAE has several user interface shortcuts to enable fast annotation and correction of entities. Users can extend an entity annotation by highlighting text that begins within an entity annotation and goes beyond the entity. Inversely, users can also shorten entity annotations by highlighting text that begins outside an entity span and finishes within an entity, thereby removing the selected text from the entity annotation. We extended this functionality to work for the new subspan system so that it would extend the appropriate first or last subspan in an entity outwards, or would shorten or even remove subspans that are highlighted. We further added user interface tweaks so that when a user selected a subspan, it would select all the subspans for the entity. Finally, we implemented export functionality so that the new subspans would be correctly stored in the PubAnnotation format with a list of spans for those entities with multiple subspans.
The code for this paper is available at https://github.com/jakelever/textae.
We first tested to check that all existing functionality of TextAE remained operational. We confirmed that the new subspan model was able to load data containing non-contiguous entities and annotations with non-contiguous entities could be saved correctly to the PubAnnotation format. Furthermore, we tested that all existing functionality, including relation annotation, worked correctly with non-contiguous entity annotations.
We quantified the user interactions required to annotate non-contiguous entities. With this new interface, the user needs to annotate a single span (1 click), enable the Add subspan mode (1 click), add a new subspan (1 click), and disable the Add subspan mode (1 click). With only four clicks, we have drastically reduced the user effort, compared to the 10 clicks required previously, and no longer require the user to switch annotation modes within TextAE. Furthermore, the output is visually clearer. This performance is similar to the Catma tool, which requires four clicks to annotate a non-contiguous entity (1 to activate the discontinuous mode, 2 to select the two spans and 1 to select the entity type). And it is marginally easier than BRAT which takes five clicks (2 to annotate the first entity, 1 to edit the entity, 1 to select Add Frag and 1 to select the new span).
While non-contiguous entities initially seem like a limited problem for text annotation, we note that two other BLAH 6 hackathon projects requested this functionality during the event: a project working on annotations from the recent BioNLP Shared Task [10] and a project focused on Medical Device Indication annotation. To understand the scale of this problem, we quantified the number of non-contiguous entities that appear in lists, as shown in the CancerMine examples in Table 1. We focussed on this format as these can be extracted using a modified dictionary matching method.
We used the PubTator Central resource [11] as it provides text-level entity annotations of a very large set of biomedical publications and also a rough set of synonyms for different entity types. The annotations provide locations of biomedical entities that may be the final element in a list. For example, the phrase ŌĆ£prostate, skin and lung cancerŌĆØ would only likely be tagged for ŌĆ£lung cancerŌĆØ in PubTator. We aimed to retrieve other entities from these lists using the set of synonyms from PubTator Central, so that ŌĆ£prostate cancerŌĆØ and ŌĆ£skin cancerŌĆØ would be extracted from the example phrase. We used a simple rule-based system that identified candidate lists by searching for tagged biomedical entities that follow the word ŌĆ£and.ŌĆØ We then searched the preceding words in the candidate list and attempted word substitutions with the final term to find terms that were in the lexicon.
Across the 30,044,935 abstracts and 2,485,641 full-text papers that were minable, we find 3,269,632 potential mentions of non-contiguous entities in the example list format. We manually reviewed 100 of them to understand the error profile and found that 42% were true positives. The main errors were caused by spurious mistakes in the lexicon and a more conservative lexicon would likely improve precision but may affect overall recall. Nevertheless, this initial result suggests that many biomedical entities are described in the list form that would be missed with most current methods. While there are considerable false-positive dues to the dictionary matching method, we would argue that this will only be a fraction of non-contiguous entities across the biomedical literature as we examine only one type of linguistic structure that could contain non-contiguous entities.
Fig. 3 shows an overview of the results from the literature analysis. Lists appear more in full-text papers than in abstracts even when taking account of the substantially larger number of abstracts than full-text articles in the corpus. They can even appear in the article title. Furthermore, disease has substantially more non-contiguous entities, likely due to the larger number of multiple word terms in that lexicon (837,390 compared to 103,427 for genes for example).
This analysis strongly suggests that non-contiguous are a substantial problem in biomedical text mining and that methods that ignore them will be missing large amounts of potential extracted information. We hope our contribution to an annotation tool that could help visualize and annotate these problematic entities may take a step towards new methods to identify them.
Notes
Acknowledgments
The authors would like to thank the funders of the Biomedical Linked Annotation Hackathon series.
Fig.┬Ā1.
Illustration of three entity annotations including two non-contiguous represented using the older chaining model which is cumbersome to annotate and visually cluttered.
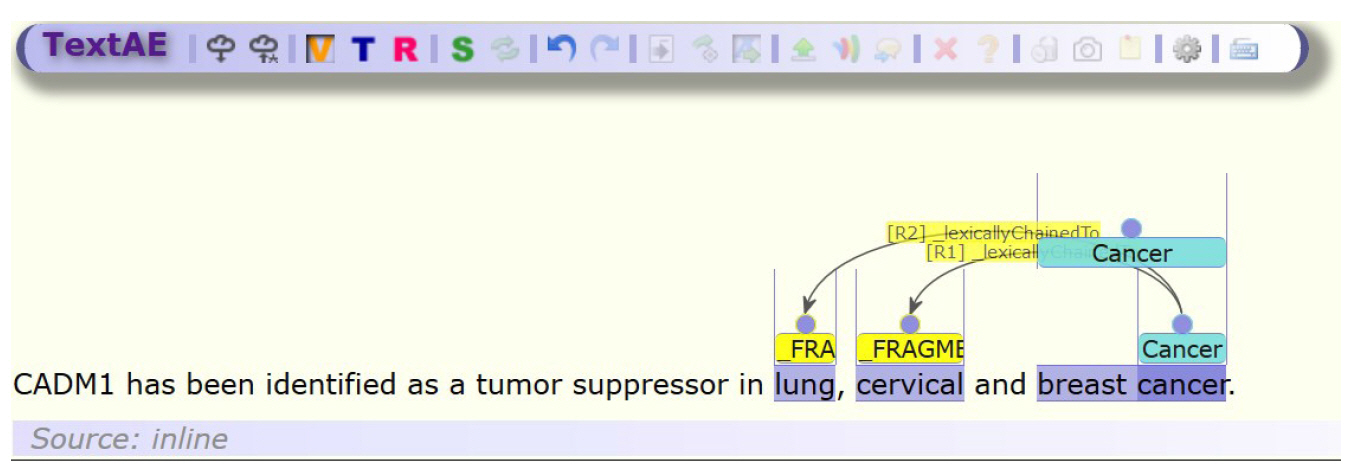
Fig.┬Ā2.
The user interface with the new Add subspan button in the toolbar (highlighted by red arrow) and non-contiguous entities annotated as part of three relationships.

Table┬Ā1.
Examples of non-contiguous entities from different public text mining datasets
Table┬Ā2.
An analysis of the annotation tools reviewed in Neves and SevaŌĆÖs study [4] for their capabilities to annotate non-contiguous entities
| Tool | URL | Can run? | Supports entity annotation? | Support non-contiguous entities? |
|---|---|---|---|---|
| BioQRator | http://www.bioqrator.org | Y | Y | N |
| brat | http://brat.nlplab.org | Y | Y | Y |
| Catma | https://catma.de | Y | Y | Y |
| Djangology | https://sourceforge.net/projects/djangology | Y | Y | N |
| ezTag | https://eztag.bioqrator.org | Y | Y | N |
| FLAT | https://github.com/proycon/flat | Y | Y | N |
| LightTag | https://www.lighttag.io | Y | Y | N |
| MAT | http://mat-annotation.sourceforge.net | Y | Y | N |
| MyMiner | http://myminer.armi.monash.edu.au | Y | Y | N |
| PDFAnno | https://github.com/paperai/pdfanno | N | - | - |
| prodigy | https://prodi.gy/ | Y | Y | N |
| tagtog | https://www.tagtog.net/ | Y | Y | N |
| WAT-SL | https://github.com/webis-de/wat | Y | N | - |
| WebAnno | https://webanno.github.io | Y | Y | N |
References
1. Leaman R, Gonzalez G. BANNER: an executable survey of advances in biomedical named entity recognition. Pac Symp Biocomput 2008;652ŌĆō633.


2. Leaman R, Wei CH, Lu Z. tmChem: a high performance approach for chemical named entity recognition and normalization. J Cheminform 2015;7:S3.




3. Leaman R, Islamaj Dogan R, Lu Z. DNorm: disease name normalization with pairwise learning to rank. Bioinformatics 2013;29:2909ŌĆō2917.




4. Neves M, Seva J. An extensive review of tools for manual annotation of documents. Brief Bioinform 2019 Dec 15 [Epub]. https://doi.org/10.1093/bib/bbz130.


5. Stenetorp P, Pyysalo S, Topic G, Ohta T, Ananiadou S, Tsujii J. BRAT: a web-based tool for NLP-assisted text annotation. In: Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics (Segond F, ed.), 2012 Apr 23-27, Avignon, France. Stroudsburg: Association for Computational Linguistics, 2012. pp. 102-107.
6. Papazian F, Bossy R, Nedellec C. AlvisAE: a collaborative Web text annotation editor for knowledge acquisition. In: Proceedings of the Sixth Linguistic Annotation Workshop (Ide N, Xia F, eds.), 2012 Jul 12-13, Jeju, Korea. Stroudsburg: Association for Computational Linguistics, 2012. pp. 149-152.
7. Kim JD, Wang Y. PubAnnotation: a persistent and sharable corpus and annotation repository. In: Proceedings of the 2012 Workshop on Biomedical Natural Language Processing (Cohen KB, Demner-Fushman D, Ananiadou S, Webber B, Tshujii J, Pestian J, eds.), 2012 Jun 3-8, Montreal, Canada. Stroudsburg: Association for Computational Linguistics, 2012. pp. 202-205.
8. Lever J, Barbarino JM, Gong L, Huddart R, Sangkuhl K, Whaley R, et al. PGxMine: text mining for curation of PharmGKB. Pac Symp Biocomput 2020;25:611ŌĆō622.



9. Lever J, Zhao EY, Grewal J, Jones MR, Jones SJ. CancerMine: a literature-mined resource for drivers, oncogenes and tumor suppressors in cancer. Nat Methods 2019;16:505ŌĆō507.



10. Bossy R, Deleger L, Chaix E, Ba M, Nedellec C. Bacteria Biotope at BioNLP Open Shared Tasks 2019. In: Proceedings of The 5th Workshop on BioNLP Open Shared Tasks (Kim JD, Nedellec C, Bossy R, Deleger L, eds.), 2019 Nov 4, Hong Kong. Stroudsburg: Association for Computational Linguistics, 2019. pp. 121-131.

- TOOLS
- Related articles in GNI