 |
 |
- Search
| Genomics Inform > Volume 16(4); 2018 > Article |
|
Abstract
Coronary artery disease (CAD) is one of the leading causes of death and disability all around the world. Recent studies have revealed that aberrantly regulated long non-coding RNA (lncRNA) as one of the main classes of cellular transcript plays a key regulatory role in transcriptional and epigenetic pathways. Recent reports have demonstrated that circulating lncRNAs in the blood can be potential biomarkers for CAD. HOTAIR is one of the most cited lncRNAs with a critical role in the initiation and progression of the gene expression regulation. Recent research on the role of the HOTAIR in cardiovascular disease lays the basis for the development of new studies considering this lncRNA as a potential biomarker and therapeutic target in CAD. In this study, we aimed to compare the expression of HOTAIR lncRNA in the blood samples of patients with CAD and control samples. The expression level was examined by semi-quantitative reverse transcriptase polymerase chain reaction technique. Our data shows that expression of HOTAIR is up-regulated in blood samples of patients with CAD.
Cardiovascular disease (CVD) is one of the leading causes of death in the world and include a group of disorders that occur in the heart or the arteries [1]. Coronary artery disease (CAD) as a most common type is caused by atherosclerosis that includes stable angina, unstable angina, myocardial infarction, and sudden cardiac death. Atherosclerosis can be created over time. It is considered a high-risk factor that could be able to restrict blood flow to the heart by increasing arterial blockage [2, 3]. Despite survival improvement in patients with CVD, specific diagnostic biomarkers have not been fully determined, and such therapies do not fully address the underlying cause.
Specific regulatory networks are required to coordinate development, physiological response, and environmental adaption of heart function that these regulatory networks are composed of pre-mediated transcriptional control and signaling pathway [4]. More recent studies have revealed that how specific physiological and pathological processes possessed by a highly diverse group of regulatory non-coding RNAs (ncRNAs) through transcriptional and epigenetic changes [5, 6]. The ncRNAs make up the majority (98%) of the transcriptome, and different classes of this regulatory RNA are discovered. They are classified based on their length and function. Long non-coding RNAs (lncRNAs) as a major group of regulatory ncRNA are longer than 200 nt in size [7, 8]. It has been clarified that some lncRNAs in the plasma were expressed differentially in CVDs, and aberrant changes of them as a new diagnostic biomarker were involved in the development and physiological function of heart disorders [9, 10]. Recent studies have depicted the important role of HOTAIR (HOX antisense intergenic RNA) in the acute myocardial infarction, vascular inflammation, and age-related CVDs but research on the role of lncRNAs in CAD is still in its infancy [11–13].
Among the lncRNAs, HOTAIR is an RNA that illustrates the importance of lncRNAs in different diseases and cancers [14, 15]. It is transcribed from the HOXC cluster and represses transcription of HOXD gene as well as different other genes through targeting the histone modifications on desired genes [16, 17].
Analyzing high-throughput data characterized the expression of lncRNA under the normal physiological condition as well as disease status. It shows that several lncRNAs are regulated during myocardial infarction and heart failure while some of them control hypertrophy [18]. Different studies indicate the potential role of HOTAIR in CVDs. Cai et al. [19] found a number of lncRNAs that were differentially expressed in circulating peripheral blood monocytes and plasma from 15 CAD patients and 15 control subjects. In their supplementary data, it was shown that HOTAIR is one of the lncRNAs that is up-regulated in circulating peripheral blood monocytes (p = 0.0054855, fold change = 2.0744882) and plasma (p = 0.002561755, fold change = 8.213123) in patients with CAD compared to non-CAD patients. Based on this, we aimed to further analyze the expression level of HOTAIR in patients with CAD compared to non-CAD patients by semi-quantitative reverse transcriptase polymerase chain reaction. Our data can provide new evidence for the potential role of HOTAIR in coronary artery disease.
CAD was confirmed by coronary angiography. The samples were composed of 20 CAD patients and 20 control individuals from Imam Khomeini Hospital of Ahvaz from 2016 to 2017 and sample’s characteristics are provided in Table 1. The informed consent was obtained from all subjects. The severity of coronary artery narrowing estimated by quantitative coronary angiography to avoid inter-observer and intera-observer variability and stenosis greater than 50% luminal diameter considered as significant CAD. On the other side control group have chosen from persons with completely normal coronary artery angiogram without obvious plaque or narrowing. Coronary artery calcification grade 2 (easily recognized) and 3 (recognized in more than half of one coronary artery) were included and grade 1 calcification (difficult to recognize) excluded to avoid controversy. None of the patients had grade 4 calcification. The process of the study was approved by Shahid Chamran University of Ahvaz, Iran.
Peripheral blood mononuclear cells (PBMC) were isolated from the peripheral blood of the patients by Ficoll according to the protocol. Total RNA was isolated from PBMC samples using RNX-plus reagent (Cinnagen, Tehran, Iran) following the manufacturer’s RNA isolation protocol. The total RNA yield was determined by gel electrophoresis and spectro-photometry. RNAs were treated with DNase (SinaClone, Tehran, Iran) in order to eliminate any DNA contamination. For real-time quantitative reverse transcription PCR (qRT-PCR), RNA was reverse transcribed to cDNA by using the reverse transcriptase enzyme. cDNA was synthesized by oligo dT and random hexamer primer in 10 μL reaction from 5 μg RNA based on manufacturer’s instructions.
Reaction mixture for PCR (20 μL) containing 1 μL cDNA template, 500 nM each of sense and antisense primers that amplified as follows: initial denaturation at 95°C for 5 min, and amplification by 32 cycles of denaturation at 95°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 30 s followed by 5 min final extension. The band intensity of PCR product was verified by electrophoresis on the agarose gel. Agarose gel stained with safe stain and band density was quantified by UV software.
The primer sequences are as following: (forward, GTCGGAGTCAACGGATTTG; reverse, TGGGTGGAATCA TATTGGA) for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and (forward, AGAAAAAGCAACCACGAAGC; reverse, AAACCTCTGTCTGTGAGTGCC) for HOTAIR.
To examine the significance of the HOTAIR expression in CVD patients, we tested 20 blood samples from CVD patients and 20 control samples. The mean age of patients was 53.95 years at the time of CAD diagnosis and control samples average ages was 50 years old.
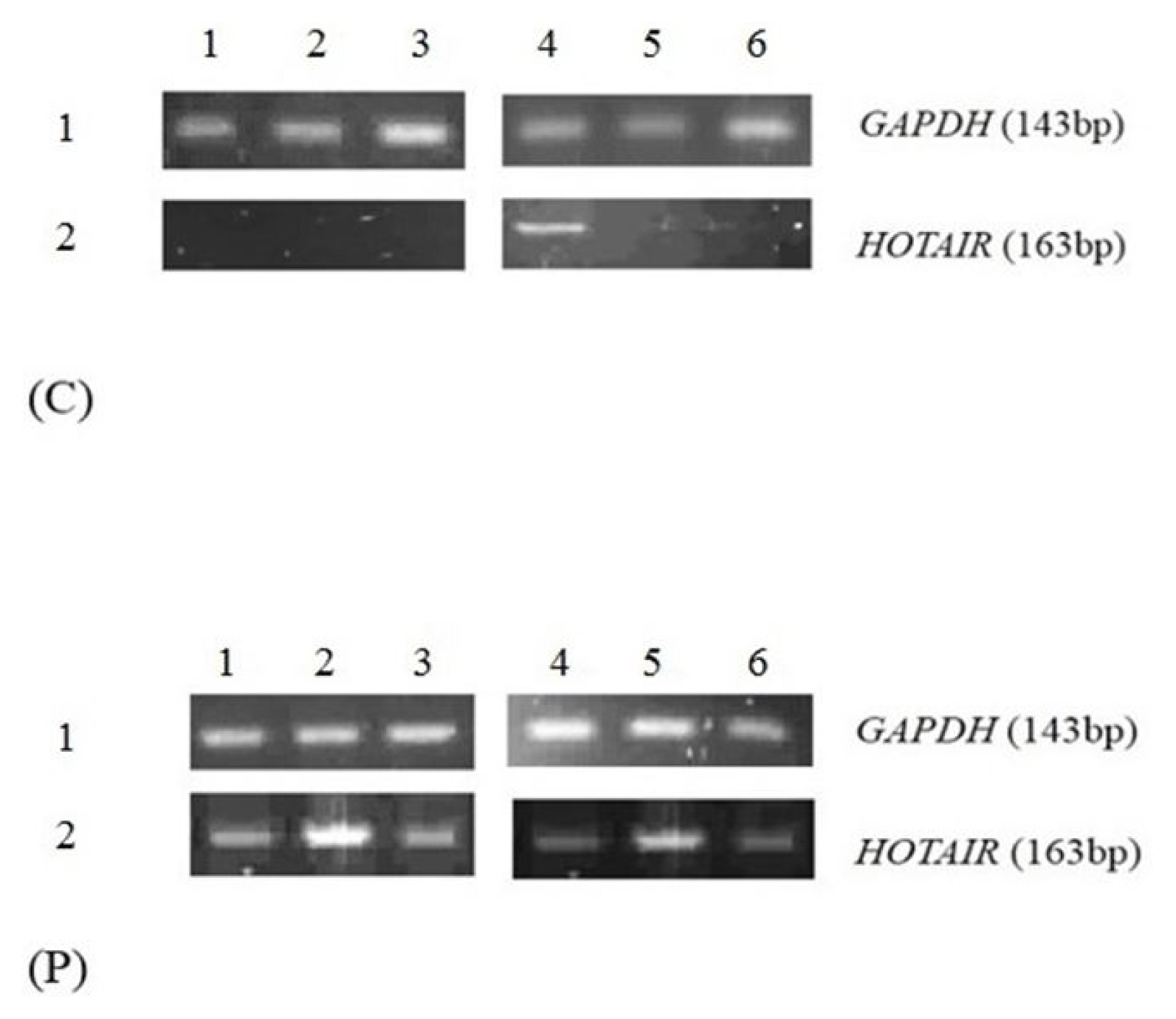
Size of products for GAPDH and HOTAIR were controlled on 2% agarose gel. The 143-bp PCR product attributed to amplified GAPDH fragment was visualized in all the control and patient’s samples while 19 out of 20 of case samples (95%) demonstrated HOTAIR PCR product. However, only 3 normal samples showed the band for HOTAIR on the agarose gel. The results indicated that the expression level of HOTAIR was higher in CAD patients (Fig. 1) compared to non-CAD patients. Unpaired student t-test showed that the relative expression of HOTAIR in CVD patient’s blood sample was significantly higher than the controls (Fig. 2) (fold change, 4.710; p < 0.0001). Also, HOTAIR expression in CVD patient and control samples were analyzed based on their age and gender. We categorized HOTAIR lncRNA expression based on gender and age. There were no significant differences of HOTAIR expression between groups.
CAD as one of the most cardiovascular disorders is the main cause of myocardial infarction [20]. Formation of fatty steaks and atherosclerotic plaques lead to the development of atherosclerosis [21, 22]. Moreover, different recent and important discoveries have demonstrated that increasing number of ncRNAs including lncRNAs and miRNAs have an effect on different diseases such as CVDs [23, 24]. Discovering the function of several lncRNAs in endothelial cells, smooth muscle cells, macrophages, vascular, inflammation, and metabolism have shown that these RNAs can be involved in the progression of atherosclerosis and CAD [25, 26]. Also, aberrantly expression of these ncRNAs in CVD have an effect on the normal physiological condition. So, it seems that circulating lncRNAs have potential application in diagnosis and treatment of CVD [27, 28].
Aberrant expression of the HOTAIR has been examined in different diseases [29]. Querying the supplementary data derived from Cai et al.’s study [19] shows that HOTAIR is up regulated in CAD patients compared to non-CAD patients. The results have been achieved by a high throughput study. It motivated us to examine the HOTAIR expression in CAD patients compared to non-CAD patients by qRT-PCR.
Here, we found that the expression levels of HOTAIR were significantly up-regulated in patient’s blood samples compared with normal ones. The results of this study demonstrate a potential role of the HOTAIR in CAD.
Recently, HOTAIR has been implicated in CVDs, especially in aortic valve calcification. Rinn et al. [30] emphasized that HOTAIR serves as a scaffold for two distinct histone modification complexes. Furthermore, Carrion et al. [31] found that HOTAIR levels are decreased in response to Leaflet stretch of aortic valve interstitial cells (AVICs). Based on the previous studies, more than half of the calcific aortic valve display bicuspid morphology symptoms. Human AVICs that exposed to stretch have a lower level of HOTAIR. Reducing the HOTAIR expression level via siRNA was followed by elevated expression of ALPL and BMP2 genes which are related to calcification [31]. Although Greco et al. [32] identified HOTAIR as one of the nine lncRNAs that are deregulated in heart failure [32], our results are inconsistent with the data presented by Cai et al. [19]. Our aim in this study is to emphasis on the enhanced expression of circulating HOTAIR in CAD patients compared to non-CAD patients for the first time. Altogether, the current study provides a viewpoint for further researches. More studies are required to approve the significance of HOTAIR and related genes as molecules involved in CAD. More experimental studies may reveal circulating lncRNAs as biomarkers or therapeutic targets in CVDs.
Notes
Fig. 1
Reverse transcription polymerase chain reaction analysis of the HOTAIR and GAPDH expression in the blood samples collected from coronary artery disease patients (p) and normal (c).
References
1. Heitzer T, Schlinzig T, Krohn K, Meinertz T, Munzel T. Endothelial dysfunction, oxidative stress, and risk of cardiovascular events in patients with coronary artery disease. Circulation 2001;104:2673–2678.


2. Wong ND. Epidemiological studies of CHD and the evolution of preventive cardiology. Nat Rev Cardiol 2014;11:276–289.



3. GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015;385:117–171.


4. Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science 2006;311:796–800.


5. Gangwar RS, Rajagopalan S, Natarajan R, Deiuliis JA. Noncoding RNAs in cardiovascular disease: pathological relevance and emerging role as biomarkers and therapeutics. Am J Hypertens 2018;31:150–165.



6. Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci 2014;39:35–43.


7. ENCODE Project Consortium, Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447:799–816.




8. Hajjari M, Khoshnevisan A, Shin YK. Molecular function and regulation of long non-coding RNAs: paradigms with potential roles in cancer. Tumour Biol 2014;35:10645–10663.


9. Ounzain S, Burdet F, Ibberson M, Pedrazzini T. Discovery and functional characterization of cardiovascular long noncoding RNAs. J Mol Cell Cardiol 2015;89:17–26.


10. Ma Y, Ma W, Huang L, Feng D, Cai B. Long non-coding RNAs, a new important regulator of cardiovascular physiology and pathology. Int J Cardiol 2015;188:105–110.


11. Gao L, Liu Y, Guo S, Yao R, Wu L, Xiao L, et al. Circulating long noncoding RNA HOTAIR is an essential mediator of acute myocardial infarction. Cell Physiol Biochem 2017;44:1497–1508.


12. Haemmig S, Simion V, Feinberg MW. Long non-coding RNAs in vascular inflammation. Front Cardiovasc Med 2018;5:22.



13. Viereck J, Thum T. Circulating Noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ Res 2017;120:381–399.


14. Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics 2016;14:42–54.



15. Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med 2015;12:1–9.


16. Cao J. The functional role of long non-coding RNAs and epigenetics. Biol Proced Online 2014;16:11.




17. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010;329:689–693.



18. Rizki G, Boyer LA. Lncing epigenetic control of transcription to cardiovascular development and disease. Circ Res 2015;117:192–206.


19. Cai Y, Yang Y, Chen X, Wu G, Zhang X, Liu Y, et al. Circulating 'lncRNA OTTHUMT00000387022' from monocytes as a novel biomarker for coronary artery disease. Cardiovasc Res 2016;112:714–724.



20. Li D, Chen G, Yang J, Fan X, Gong Y, Xu G, et al. Transcriptome analysis reveals distinct patterns of long noncoding RNAs in heart and plasma of mice with heart failure. PLoS One 2013;8:e77938.



21. Kadar A, Glasz T. Development of atherosclerosis and plaque biology. Cardiovasc Surg 2001;9:109–121.


22. Libby P. Vascular biology of atherosclerosis: overview and state of the art. Am J Cardiol 2003;91:3A–6A.


23. Greco S, Gorospe M, Martelli F. Noncoding RNA in age-related cardiovascular diseases. J Mol Cell Cardiol 2015;83:142–155.



24. Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol 2010;220:126–139.


25. Liu Y, Zheng L, Wang Q, Hu YW. Emerging roles and mechanisms of long noncoding RNAs in atherosclerosis. Int J Cardiol 2017;228:570–582.


26. Zhou T, Ding JW, Wang XA, Zheng XX. Long noncoding RNAs and atherosclerosis. Atherosclerosis 2016;248:51–61.


27. Thum T, Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res 2015;116:751–762.


28. Archer K, Broskova Z, Bayoumi AS, Teoh JP, Davila A, Tang Y, et al. Long non-coding RNAs as master regulators in cardiovascular diseases. Int J Mol Sci 2015;16:23651–23667.



29. Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett 2013;339:159–166.


30. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007;129:1311–1323.



- TOOLS
-
METRICS

-
- 15 Crossref
- 0 Scopus
- 5,331 View
- 168 Download
- Related articles in GNI








