|
 |
- Search
| Genomics Inform > Volume 16(4); 2018 > Article |
|
Abstract
Long non-coding RNAs (lncRNAs) are classified as RNAs that are longer than 200 nucleotides and cannot be translated into protein. Several studies have demonstrated that lncRNAs are directly or indirectly involved in a variety of biological processes and in the regulation of gene expression. In addition, lncRNAs have important roles in many diseases including cancer. It has been shown that abnormal expression of lncRNAs is observed in several human solid tumors. Several studies have shown that many lncRNAs can function as oncogenes in cancer development through the induction of cell cycle progression, cell proliferation and invasion, anti-apoptosis, and metastasis. Oncogenic lncRNAs have the potential to become promising biomarkers and might be potent prognostic targets in cancer therapy. However, the biological and molecular mechanisms of lncRNA involvement in tumorigenesis have not yet been fully elucidated. This review summarizes studies on the regulatory and functional roles of oncogenic lncRNAs in the development and progression of various types of cancer.
Upon completion of the Human Genome Project, it was revealed that only 2% of the human genome including protein-coding genes [1]. In the past, non-coding RNAs (ncRNAs) were deemed so-called useless or ŌĆśjunkŌĆÖ DNA. But, as a result of the Encyclopedia of DNA Elements (ENCODE) project, it was found that ncRNAs have specific functions in a variety of biological processes [2]. Moreover, it has been shown that 98% of ncRNAs are involved in various cellular functions including chromatin remodeling, transcription, alternative splicing and translational regulation.
In addition, there are several types of ncRNAs including small non-coding RNAs (sncRNAs), microRNAs (miRNAs), and small nucleolar RNAs. Recently, it was reported long non-coding RNAs (lncRNAs) that were different from sncRNAs [3]. The classification standard for sncRNAs and lncRNAs is based on nucleotide (nt) length, and the length criterion for sncRNAs was less than 200 nt while the length of lncRNAs was more than 200 nt [4]. Accumulated clues have been reported that lncRNAs have important roles in many biological processes such as cell growth and differentiation, chromatin composition, and the regulation of gene expression by directly or indirectly participating in transcription and translation, indicating that lncRNAs could act as functional genes [5]. Thus, there are many efforts conducted by many oncologists to identify and analyze lncRNAs as novel functional genes responsible for tumorigenesis and cancer development. It has been revealed that some of lncRNAs act as oncogenes highly associated with the hallmarks of cancer such as cell cycle progression, anti-apoptosis, angiogenesis, and metastasis [6ŌĆō8]. In this review, we discuss the roles of lncRNAs in biological processes and describe the lncRNA-mediated signaling pathways involved in cancers, with a focus on oncogenic lncRNAs.
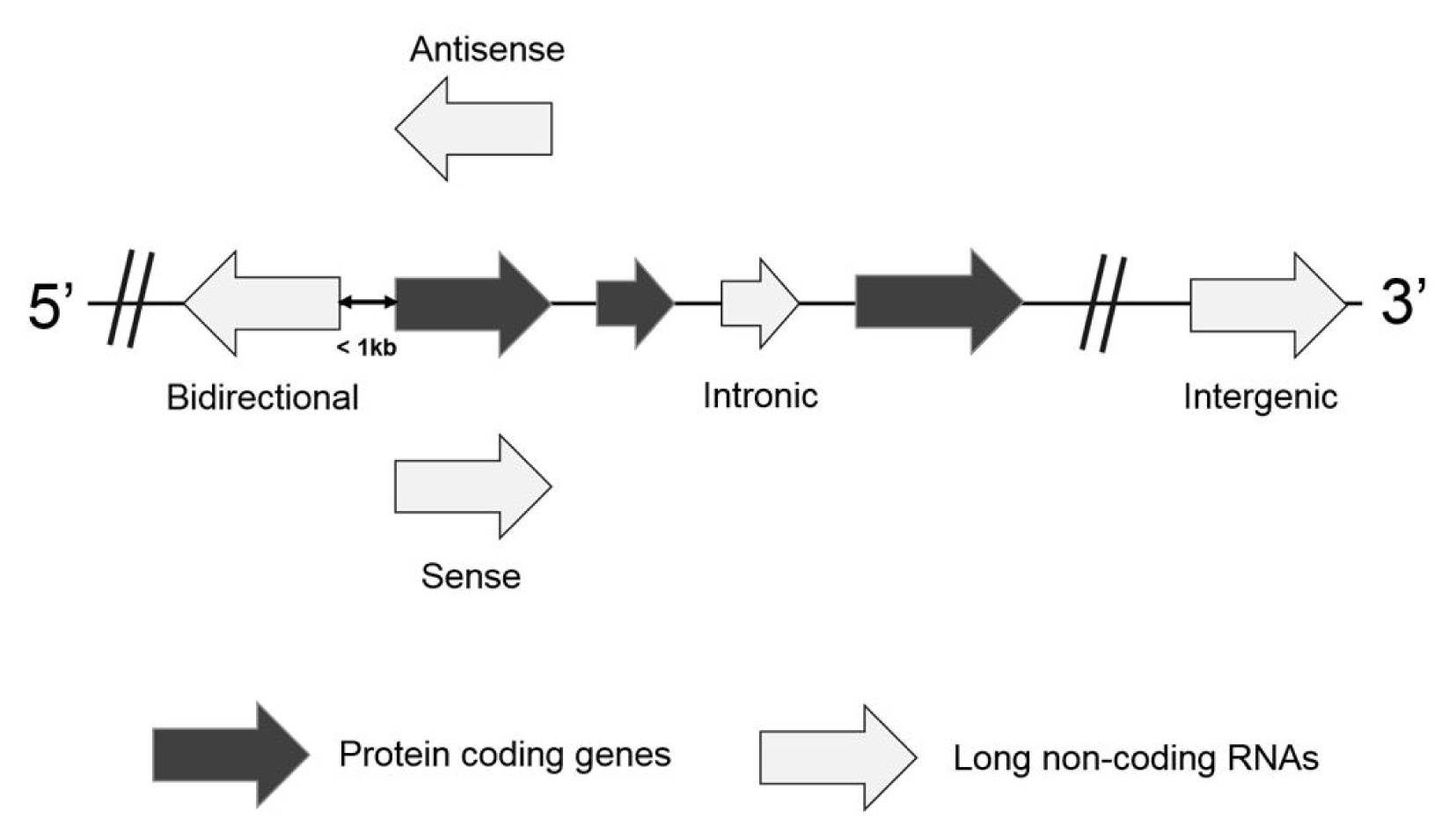
Based on the location and orientation on the chromosome, lncRNAs are subdivided as intergenic lncRNAs, intronic lncRNAs, bidirectional lncRNAs, sense overlapping lncRNAs, and antisense RNAs (Fig. 1) [9, 10]. Briefly, an intergenic lncRNA locates between two protein-coding genes more than 1 kb away from each other. An intronic lncRNA is placed on the intron region of a protein coding gene in either sense or antisense direction. A biodirectional lncRNA is oriented from the opposite direction within 1 kb of the promoter for the protein-coding gene. In particular, when a protein-coding gene is transcribed, a biodirectional lncRNA counterpart tends to be transcribed as well. It suggests that they might share regulatory factors for the transcription. A sense overlapping lncRNA overlaps with a protein-coding gene on the same DNA strand for its transcription. Since this type of lncRNA extensively lacks the open reading frame for the translation, it can be distinguished from a transcript variant of the protein-coding mRNAs. An antisense lncRNA can be transcribed from the antisense DNA strand of a protein-coding gene. Despite several types of lncRNAs, they are highly probable to have been transcribed by RNA polymerase II, capping, splicing, and polyadenylation [11]. They are also controlled by transcription activators and repressors. However, it has been reported that lncRNAs have limited coding potential.
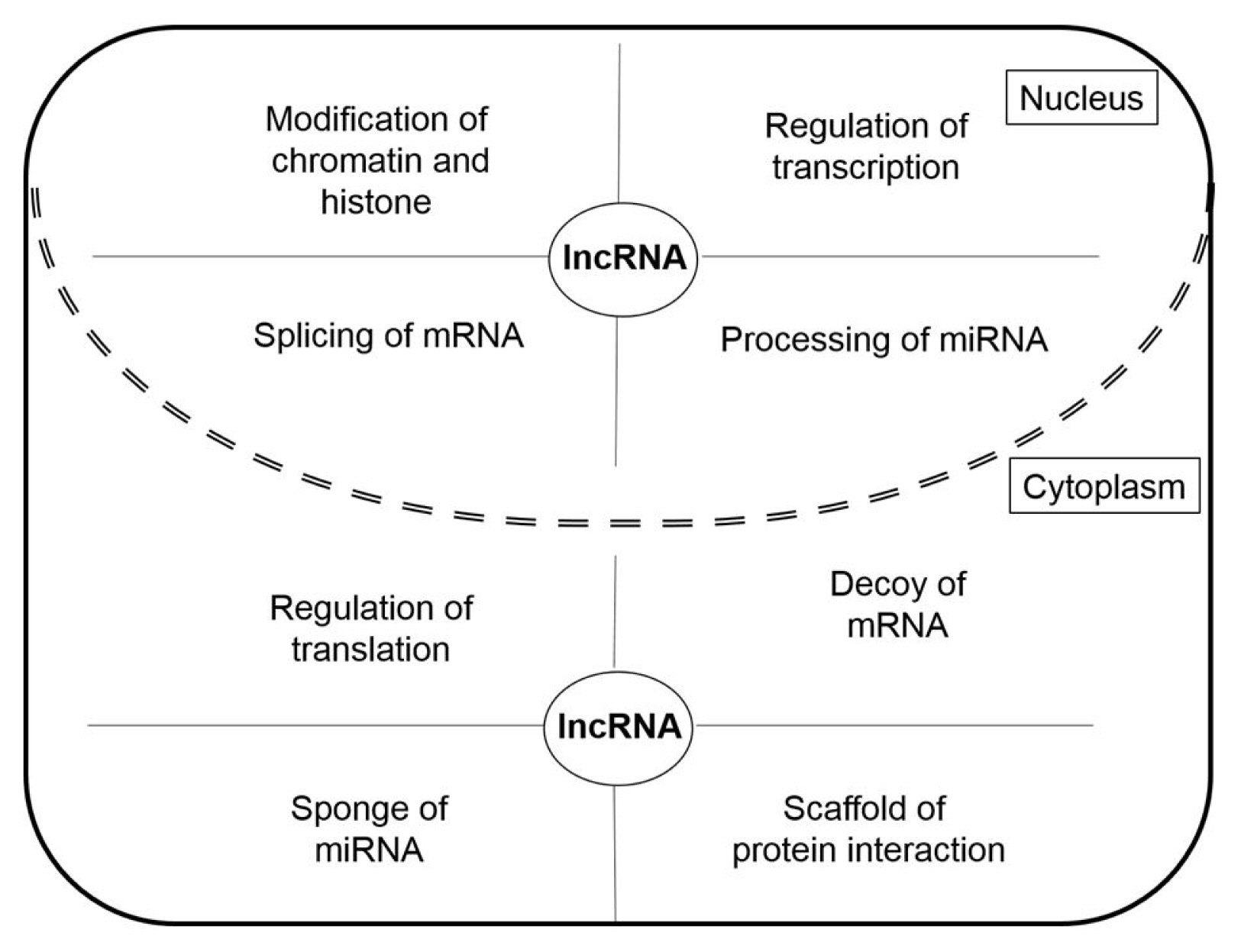
LncRNAs have different roles in nucleus and cytoplasm (Fig. 2). The lncRNAs in the nucleus function as chromatin remodeling and transcription guides and are active in the regulation of transcription factors (TFs) and TF-associated factors, as well as the regulation of pre-mRNA splicing and miRNA processing [12]. LncRNAs in the cytoplasm function as competing endogenous RNAs (ceRNAs) acting as molecular sponges for miRNAs, targeting proteins to regulate their expression, and suppressing translation by targeting mRNAs [13]. In addition to their various roles in cells, lncRNAs are also involved in various human diseases including cardiovascular diseases, autoimmune diseases, neurodegenerative disorders, and psychiatric disorders [10, 14, 15]. In particular, lncRNAs are reported to be closely associated with tumorigenesis [8]. Many lncRNAs act as oncogenic genes in the development of cancer through the regulation of metastasis, cell death suppression, and cell growth, as well they affect the DNA damage response through their relationships with various genes [16]. In the next section, we demonstrate the functions of cancer-related oncogenic lncRNAs in lung cancer, colorectal cancer, glioblastoma, and breast cancer.
Lung cancer is the leading cause of cancer-related deaths in the world [17]. Eighty-five percent of lung cancer is classified as non-small cell lung cancer (NSCLC) and the remaining 15% is categorized as small-cell lung cancer [18]. Approximately 70% of NSCLC patients are diagnosed at an advanced cancer metastatic state and their 5-year probability of survival is approximately 14% [19]. Various lncRNAs have been implicated in NSCLC including, PVT1, LINC00152, HOTAIR, H19, UPAT, HOXD-AS1, LINC00339, and MLAT (Table 1).
The lncRNA plasmacytoma variant translocation 1 (PVT1) was first described in 2013 in human colorectal cancer. PVT1, located at 8q24, has been shown to be regulated in the biological processes of various cancer types, including liver, colon, and ovarian cancers [20ŌĆō22]. Recent studies have shown that PVT1 expression is positively associated with NSCLC aggressiveness [23]. PVT1 promotes cell growth, cancer cell invasion and anti-apoptosis by acting as a ceRNA to sponge miR-195 or miR-497 [24, 25]. In addition, PVT1 upregulates matrix metalloproteinase 9 (MMP9) by acting as a ceRNA to suppress miR-200a/b activity, thereby increasing cancer cell invasion capacity through increased MMP9 expression [25]. The oncogenic roles of PVT1 could be supported by the bioinformatics analyses based on the Oncomine database (http://www.oncomine.org/) demonstrating that PVT1 expression was significantly upregulated in lung adenocarcinomas (2.232- and 4.293-fold change) compared to the corresponding normal tissues (Fig. 3) [26, 27].
Overexpression of long intergenic ncRNA 152 (LINC00152) promotes the development of NSCLC. One study reported that LINC00152 can upregulate the expression of epidermal growth factor receptor (EGFR), which acts to upregulate tumor cell proliferation by increasing the activation of the phosphoinositide 3-kinase (PI3K)/Akt pathway [28]. Moreover, increased expression of LINC00152 increases the protein levels of fibronectin and vimentin and decreases the protein levels of p21, indicating that the expression of LINC00152 is associated with cancer cell invasion and anti-apoptosis. This study was supported by another effort conducting an RNA immunoprecipitation assay with LINC00152 and EGFR [29]. It revealed that LINC00152 could bind to EGFR (the potential peptide sequence of EGFR for binding target: MHLPSPTDSNFYR) and might allow EGFR to be phosphorylated for its activation. In addition, an increase in LINC00152 expression can decrease the level of tumor suppressor interleukin-24 by binding to enhancer of zeste homolog 2 (EZH2), which was supported by an RNA immunoprecipitation assay [30]. It indicated that LINC00152 expression can promote cell cycle progression and cell proliferation.
Colorectal cancer (CRC) is the third leading cause of morbidity and, after lung cancer, the second leading cause of cancer-related deaths in the world [17]. In recent decades, many studies of CRC have revealed several tumor-associated molecular markers, such as PI3K, BRAF, and KRAS [31]. Despite advances in treatment therapies, survival rates have remained less than 50% due to insufficient therapeutic tolerance [32]. The lncRNAs that have been associated with CRC include SNHG1, GAPLINC, TP73-AS1, CASC15, SOX21-AS1, ZEB1-AS1, DLEU1, and LINC00174 (Table 2).
Small nucleolar RNA host gene 1 (SNHG1) is reported to improve cancer cell proliferation and invasion in CRC. In addition, SNHG1 can function as a sponge of miR-145 leading to block the expression of miR-145 [33], which has targets associated with tumorigenesis including insulin-like growth factor 1 receptor and p21-activated kinase 4 [34]. Moreover, expression of SNHG1 promotes activation of the Wnt/╬▓-catenin signaling pathway by direct binding to and inhibiting miR-302/372/373/520, leading to CRC cell proliferation [35, 36]. The oncogenic roles of SNHG1 could be supported by the bioinformatics analyses based on the colon and rectum adenocarcinoma gene expression data conducted by The Cancer Genome Atlas deposited in Oncomine database demonstrating that SNHG1 expression was significantly upregulated in cecum adenocarcinomas (2.697-fold change), rectal adenocarcinomas (3.333-fold change), colon adenocarcinomas (2.806-fold change), and colon mucinous adenocarcinomas (2.241-fold change) compared to the corresponding normal tissues (Fig. 4).
The expression of gastric adenocarcinoma predictive long intergenic non-coding (GAPLINC) is upregulated in CRC. GAPLINC can directly bind to PTB-associated splicing factor and non-POU-domain-containing octamer-binding protein supported by RNA pull-down assays, which lead to upregulation of SNAI2 expression and subsequent induction of cancer cell invasion [37]. In addition, GAPLINCR can downregulate miR-34a by acting as a ceRNA, resulting in increased levels of a proto-oncogene c-MET (a target of miR-34a), which is associated with invasion by and migration of cancer cells [38].
Glioma, which is present in more than 30% of brain cancer patients, is one of the cancers with the poorest prognosis [39]. Despite the advancements in technology and therapeutic approaches for glioma treatment, its prognosis is poor and recurrence of glioma progression might be unavoidable [40]. Gliomas are sub-divided into grades IIŌĆōIV with glioblastomas (GBM), with the highest grade, having a high risk of invasion and recurrence [41, 42]. Despite many studies investigating strategies for surgery, radiotherapy, and chemotherapy of GBM, the survival period for patients with GBM is short [43]. The lncRNAs that have been associated with GBM include NEAT1, DANCR, lncHERG, SNHG7, MNX1-AS1, MCM3AP-AS1, and LINC01446 (Table 3).
The expression of nuclear paraspeckle assembly transcript 1 (NEAT1) is higher in GBM cells than in normal brain cells. NEAT1 act as a sponge of miR-449b-5p, leading to downregulation of the miRNA expression [44]. Consequently, the expression of c-Met, which has a tumorigenic role and is a molecular target of miR-449b-5p, is increased. Another study has presented that NEAT1 can also act as a ceRNA for miR-132 inhibition, which lead to increase of oncogene SOX2 expression and subsequent induction of cell proliferation, invasion, and migration in glioma cells [45]. The oncogenic roles of NEAT1 could be supported by the bioinformatics analyses based on the Oncomine database demonstrating that NEAT1 expression was significantly upregulated in GBM (5.286- and 4.887-fold change) compared to the corresponding normal brain tissues (Fig. 5) [46, 47].
Differentiation antagonizing non-protein coding RNA (DANCR) is another oncogenic lncRNA present in GBM cells. DANCR is reported to inhibit several tumor-suppressive miRNAs including miR-33b-5p, miR-1-3p, miR-206, and miR-613 by functioning as a ceRNA [48]. Inhibition of these miRs by DANCR is associated with activation of AXL/PI3K/Akt/NF-╬║B signaling. Another study has demonstrated that DANCR can activate Wnt/╬▓-catenin signaling although further study for identification of potential DANCR targets on Wnt/╬▓-catenin signaling would be required [49]. These effects of DANCR is responsible for cell proliferation, anti-apoptosis, and cell migration functions, and, as a consequence, contributes to chemoresistance in GBM therapy.
Breast cancer (BC) is the most frequently diagnosed cancer and the leading cause of cancer death in women [17]. Despite many studies into therapeutic technologies and strategies for BC in recent decades, BC remains among the leading causes of morbidity and mortality worldwide [50]. The lncRNAs associated with BC include MALAT1, CCAT2, PRLB, SUMO1P3, LINC00518, ITGB2-AS1, ARNILA, and PANDAR (Table 4).
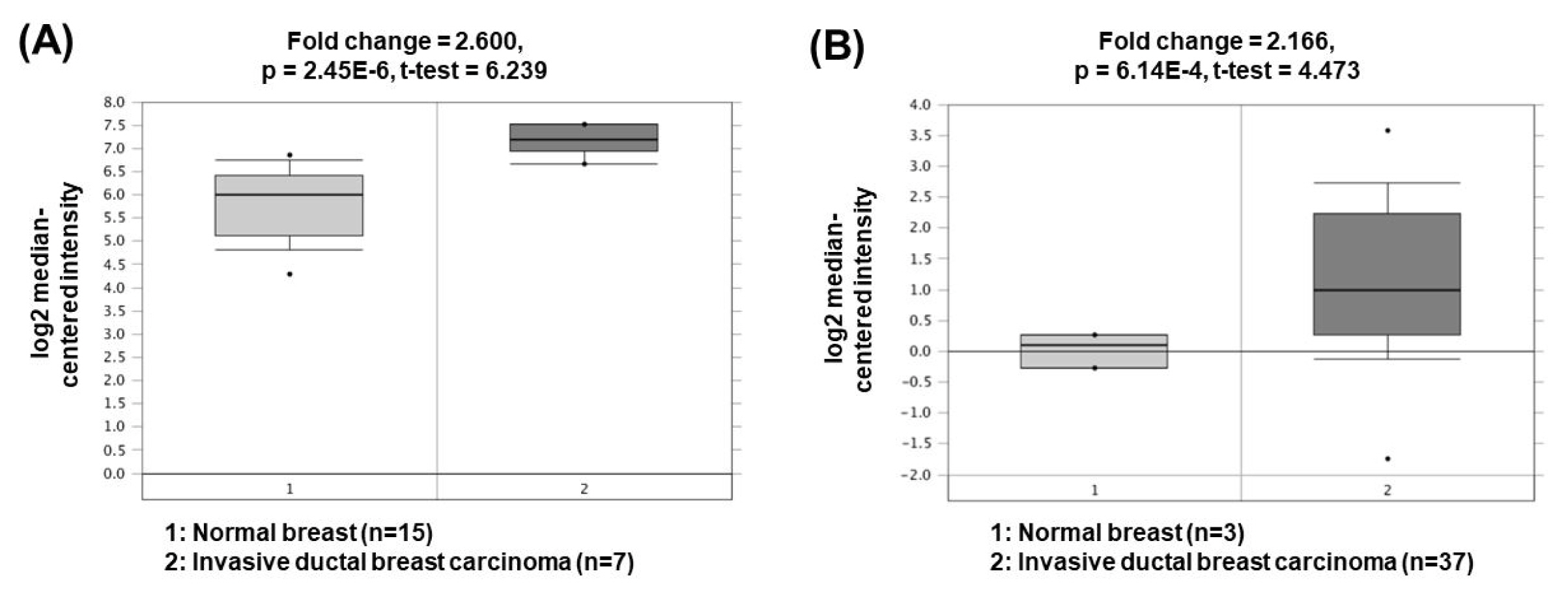
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is one of the oncogenic lncRNAs related to BC. Overexpression of MALAT1 activates cyclin-dependent kinase 4 (CDK4) by functioning as a ceRNA for inhibition of miR-124 expression [51]. Since CDK4 is a major regulator of the cell cycle and E2F1 is a downstream target of CDK4, CDK4/E2F1 signaling activated by MALAT1 is closely associated with cell cycle progression and cancer cell proliferation. Another study has presented that MALAT1 can also act as a ceRNA to sponge miR-204 [52]. Decreased miR-204 can upregulate the expression of zinc finger E-box binding homeobox 2, thereby increasing epithelial-mesenchymal transition capacity and BC cell invasion. The oncogenic roles of MALAT1 could be supported by the bioinformatics analyses based on the Oncomine database demonstrating that MALAT1 expression was significantly upregulated in invasive ductal breast carcinomas (2.600- and 2.166-fold change) compared to the corresponding normal breast tissues (Fig. 6) [53, 54].
Colon cancer-associated transcript 2 (CCAT2) is one of the lncRNAs which expression is higher in BC cells than in normal breast cells. CCAT2 can decrease tumor suppressor p15 by direct interaction with EZH2, which supported by an RNA immunoprecipitation assay [55]. Based on this effect of CCAT2, overexpressed CCAT2 can indirectly increase cell cycle-associated proteins including cyclin D1, cyclin E1, and CDK4, thereby promoting BC cell proliferation. Other study have reported that upregulated CCAT2 can induce cell growth, invation, metastasis, and anti-apoptosis in BC cells through its ability to regulate the transforming growth factor ╬▓/SMAD signaling and Wnt/╬▓-catenin signaling although specific targets of CCAT2 on both signaling pathways have remained unclear [56, 57].
Initially, ncRNAs, which do not encode proteins, were referred to as noise or junk DNA. However, interest in ncRNAs increased following the completion of the Human Genome Project. Since then, the study of ncRNAs has gradually become a central research area in RNA biology. As the amount of research on ncRNAs continues to grow, the importance of the non-protein-coding part of the human genome has been highlighted. In addition, with the revealing of several important roles of lncRNAs in vivo, interest in lncRNAs has increased. Research into the roles of lncRNAs has presented a new direction for research into tumorigenic mechanisms and cancer therapies. In general, an increase in lncRNAs expression is observed to be highly correlated to the incidence of cancer. In addition, upregulated lncRNAs contribute to the promotion of many biological processes associated with cancer development such as cell cycle progression as well as cell proliferation, invasion, metastasis, and chemoresistance. Despite the findings of many lncRNA-related studies, there are still many obstacles to fully elucidating the functions of lncRNAs. Regardless, future lncRNA studies are expected to reveal and describe the regulatory networks and molecular characteristics of human cancers. Elucidation of the relationships between oncogenic lncRNAs and tumorigenesis may be helpful for in the development of deeper insights into cancer prevention and treatment.
Notes
Fig.┬Ā3
Expression levels of PVT1 in non-small cell lung cancers. Datasets obtained from Oncomine demonstrated that lung adenocarcinomas significantly expressed high levels of PVT1 compared to the normal counterparts. (A) Fold change = 2.232, p = 6.13 ├Ś 10ŌłÆ8 [26]. (B) Fold change = 4.293, p = 1.45 ├Ś 10ŌłÆ5 [27]. PVT1, plasmacytoma variant translocation 1.
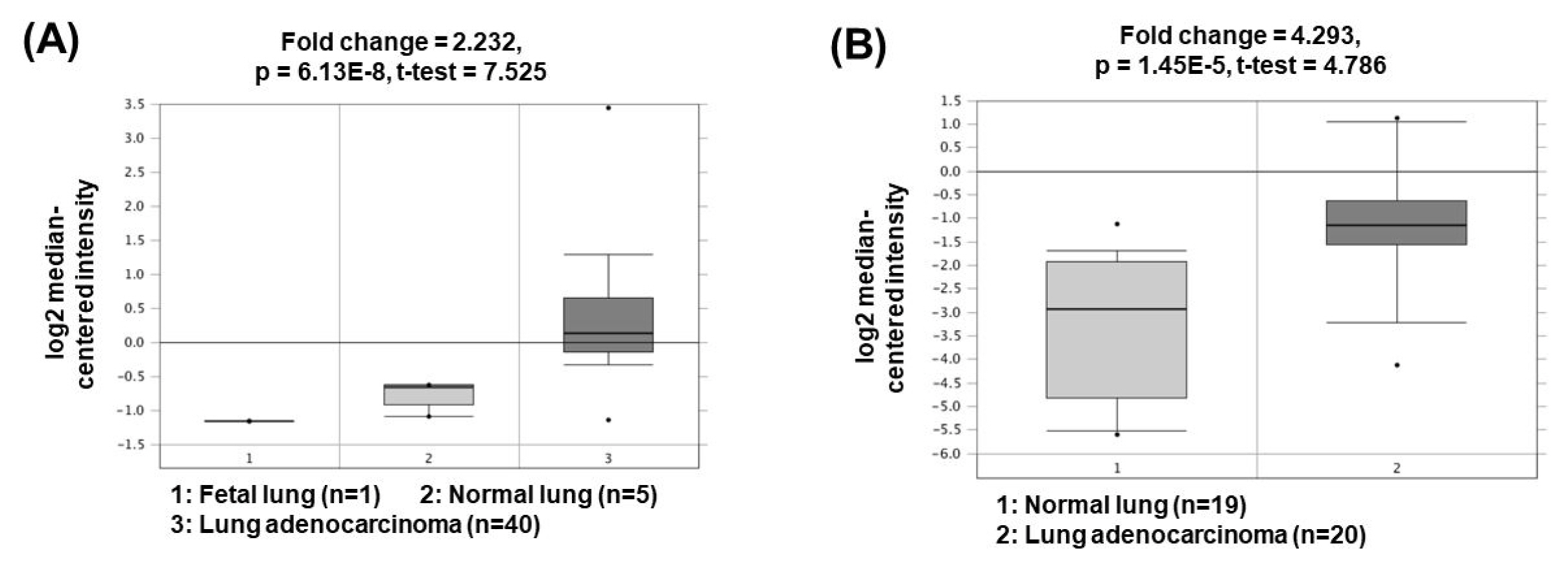
Fig.┬Ā4
Expression levels of SNHG1 in colorectal cancers. Datasets obtained from Oncomine demonstrated that cecum adenocarcinomas (A) (fold change = 2.697, p = 5.22 ├Ś 10ŌłÆ14), rectal adenocarcinomas (B) (fold change = 3.333, p = 1.08 ├Ś 10ŌłÆ21), colon adenocarcinomas (C) (fold change = 2.806, p = 2.31 ├Ś 10ŌłÆ19), and colon mucinous adenocarcinomas (D) (fold change = 2.241, p = 1.10 ├Ś 10ŌłÆ10) significantly expressed high levels of PVT1 compared to the normal counterparts. SNHG1, small nucleolar RNA host gene 1; PVT1, plasmacytoma variant translocation 1.

Fig.┬Ā5
Expression levels of NEAT1 in glioblastomas. Datasets obtained from Oncomine demonstrated that GBM significantly expressed high levels of NEAT1 compared to the normal counterparts. (A) Fold change = 5.286, p = 3.18 ├Ś 10ŌłÆ14 [46]. (B) Fold change = 4.887, p = 1.15 ├Ś 10ŌłÆ11 [47]. NEAT1, nuclear paraspeckle assembly transcript 1; GBM, glioblastomas.
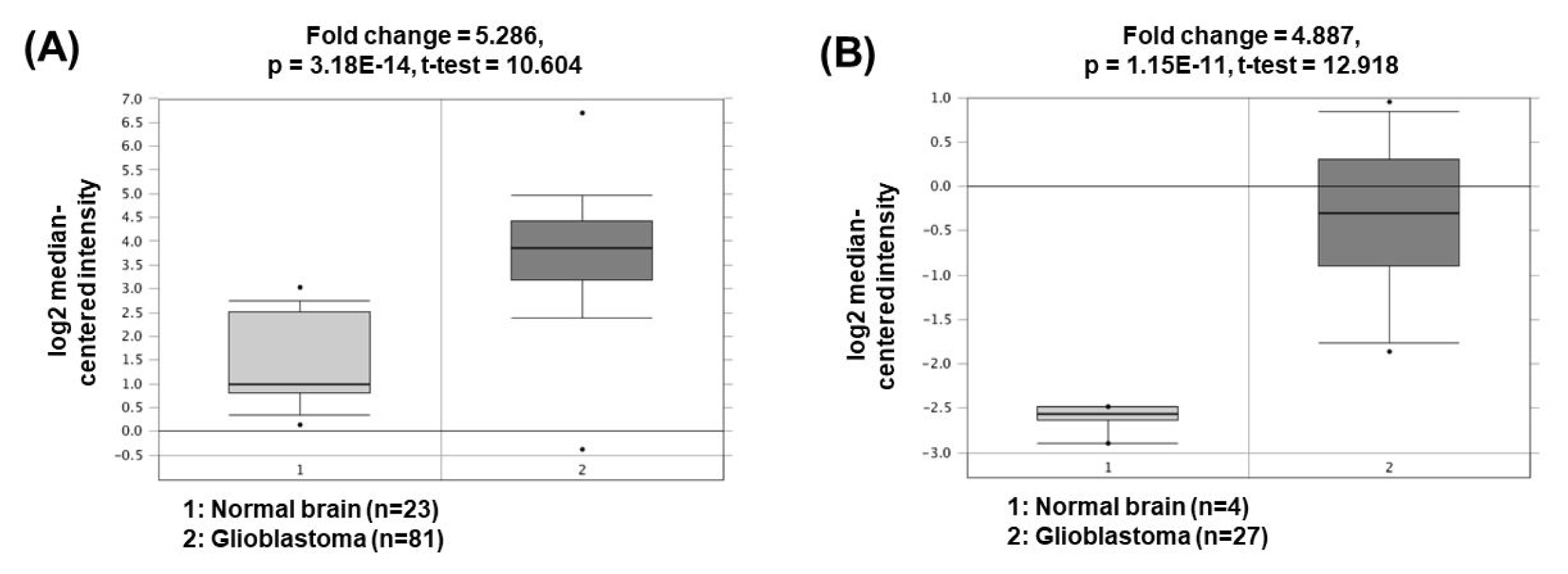
Fig.┬Ā6
Expression levels of MALAT1 in breast cancers. Datasets obtained from Oncomine demonstrated that invasive ductal breast carcinomas significantly expressed high levels of MALAT1 compared to the normal counterparts. (A) Fold change = 2.600, p = 2.45 ├Ś 10ŌłÆ6 [53]. (B) Fold change = 2.166, p = 6.14 ├Ś 10ŌłÆ4 [54]. MALAT1, metastasis-associated lung adenocarcinoma transcript 1.
Table┬Ā1
Oncogenic lncRNAs in non-small cell lung cancers
Table┬Ā2
Oncogenic lncRNAs in colorectal cancers
Table┬Ā3
Oncogenic lncRNAs in glioblastomas
Table┬Ā4
Oncogenic lncRNAs in breast cancers
References
1. International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004;431:931ŌĆō945.



2. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775ŌĆō1789.



3. Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest 2016;126:2775ŌĆō2782.



4. Zhang H, Chen Z, Wang X, Huang Z, He Z, Chen Y. Long non-coding RNA: a new player in cancer. J Hematol Oncol 2013;6:37.



5. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629ŌĆō641.


6. Kitagawa M, Kitagawa K, Kotake Y, Niida H, Ohhata T. Cell cycle regulation by long non-coding RNAs. Cell Mol Life Sci 2013;70:4785ŌĆō4794.




7. Khorshidi A, Dhaliwal P, Yang BB. Noncoding RNAs in tumor angiogenesis. Adv Exp Med Biol 2016;927:217ŌĆō241.


8. Shen XH, Qi P, Du X. Long non-coding RNAs in cancer invasion and metastasis. Mod Pathol 2015;28:4ŌĆō13.


9. Quan Z, Zheng D, Qing H. Regulatory roles of long non-coding RNAs in the central nervous system and associated neurodegenerative diseases. Front Cell Neurosci 2017;11:175.



10. Bar C, Chatterjee S, Thum T. Long noncoding RNAs in cardiovascular pathology, diagnosis, and therapy. Circulation 2016;134:1484ŌĆō1499.


11. Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev 2015;36:25ŌĆō64.



12. Nobili L, Ronchetti D, Taiana E, Neri A. Long non-coding RNAs in B-cell malignancies: a comprehensive overview. Oncotarget 2017;8:60605ŌĆō60623.



13. Hart RP, Goff LA. Long noncoding RNAs: central to nervous system development. Int J Dev Neurosci 2016;55:109ŌĆō116.



14. Wu GC, Pan HF, Leng RX, Wang DG, Li XP, Li XM, et al. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun Rev 2015;14:798ŌĆō805.


15. Gibbons A, Udawela M, Dean B. Non-coding RNA as novel players in the pathophysiology of schizophrenia. Noncoding RNA 2018;4:E11.

16. Sun J, Bie B, Zhang S, Yang J, Li Z. Long non-coding RNAs: critical players in hepatocellular carcinoma. Int J Mol Sci 2014;15:20434ŌĆō20448.



18. Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG, et al. Non-small-cell lung cancer. Lancet 2011;378:1727ŌĆō1740.


19. Alvarez M, Roman E, Santos ES, Raez LE. New targets for non-small-cell lung cancer therapy. Expert Rev Anticancer Ther 2007;7:1423ŌĆō1437.


20. Fan H, Zhu JH, Yao XQ. Knockdown of long noncoding RNA PVT1 reverses multidrug resistance in colorectal cancer cells. Mol Med Rep 2018;17:8309ŌĆō8315.



21. Yang Q, Yu Y, Sun Z, Pan Y. Long non-coding RNA PVT1 promotes cell proliferation and invasion through regulating miR-133a in ovarian cancer. Biomed Pharmacother 2018;106:61ŌĆō67.


22. Guo J, Hao C, Wang C, Li L. Long noncoding RNA PVT1 modulates hepatocellular carcinoma cell proliferation and apoptosis by recruiting EZH2. Cancer Cell Int 2018;18:98.




23. Wang M, Ma X, Zhu C, Guo L, Li Q, Liu M, et al. The prognostic value of long non coding RNAs in non small cell lung cancer: a meta-analysis. Oncotarget 2016;7:81292ŌĆō81304.



24. Guo D, Wang Y, Ren K, Han X. Knockdown of LncRNA PVT1 inhibits tumorigenesis in non-small-cell lung cancer by regulating miR-497 expression. Exp Cell Res 2018;362:172ŌĆō179.


25. Chen W, Zhu H, Yin L, Wang T, Wu J, Xu J, et al. lncRNA-PVT1 facilitates invasion through upregulation of MMP9 in non-small cell lung cancer cell. DNA Cell Biol 2017;36:787ŌĆō793.


26. Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A 2001;98:13784ŌĆō13789.



27. Stearman RS, Dwyer-Nield L, Zerbe L, Blaine SA, Chan Z, Bunn PA Jr, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol 2005;167:1763ŌĆō1775.



28. Zhang Y, Xiang C, Wang Y, Duan Y, Liu C, Jin Y, et al. lncRNA LINC00152 knockdown had effects to suppress biological activity of lung cancer via EGFR/PI3K/AKT pathway. Biomed Pharmacother 2017;94:644ŌĆō651.


29. Zhou J, Zhi X, Wang L, Wang W, Li Z, Tang J, et al. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J Exp Clin Cancer Res 2015;34:135.



30. Chen QN, Chen X, Chen ZY, Nie FQ, Wei CC, Ma HW, et al. Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression. Mol Cancer 2017;16:17.




31. Mao C, Wu XY, Yang ZY, Threapleton DE, Yuan JQ, Yu YY, et al. Concordant analysis of KRAS, BRAF, PIK3CA mutations, and PTEN expression between primary colorectal cancer and matched metastases. Sci Rep 2015;5:8065.



32. Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012;486:532ŌĆō536.




33. Tian T, Qiu R, Qiu X. SNHG1 promotes cell proliferation by acting as a sponge of miR-145 in colorectal cancer. Oncotarget 2018;9:2128ŌĆō2139.


34. Sheng N, Tan G, You W, Chen H, Gong J, Chen D, et al. MiR-145 inhibits human colorectal cancer cell migration and invasion via PAK4-dependent pathway. Cancer Med 2017;6:1331ŌĆō1340.



35. Zhu Y, Li B, Liu Z, Jiang L, Wang G, Lv M, et al. Up-regulation of lncRNA SNHG1 indicates poor prognosis and promotes cell proliferation and metastasis of colorectal cancer by activation of the Wnt/beta-catenin signaling pathway. Oncotarget 2017;8:111715ŌĆō111727.



36. Wang H, Wang G, Gao Y, Zhao C, Li X, Zhang F, et al. Lnc-SNHG1 activates the TGFBR2/SMAD3 and RAB11A/Wnt/beta-catenin pathway by sponging MiR-302/372/373/520 in invasive pituitary tumors. Cell Physiol Biochem 2018;48:1291ŌĆō1303.


37. Yang P, Chen T, Xu Z, Zhu H, Wang J, He Z. Long noncoding RNA GAPLINC promotes invasion in colorectal cancer by targeting SNAI2 through binding with PSF and NONO. Oncotarget 2016;7:42183ŌĆō42194.



38. Luo Y, Ouyang J, Zhou D, Zhong S, Wen M, Ou W, et al. Long noncoding RNA GAPLINC promotes cells migration and invasion in colorectal cancer cell by regulating miR-34a/c-MET signal pathway. Dig Dis Sci 2018;63:890ŌĆō899.



39. Liu S, Mitra R, Zhao MM, Fan W, Eischen CM, Yin F, et al. The potential roles of long noncoding RNAs (lncRNA) in glioblastoma development. Mol Cancer Ther 2016;15:2977ŌĆō2986.



40. Ohba S, Hirose Y. Current and future drug treatments for glioblastomas. Curr Med Chem 2016;23:4309ŌĆō4316.


41. Grant R, Kolb L, Moliterno J. Molecular and genetic pathways in gliomas: the future of personalized therapeutics. CNS Oncol 2014;3:123ŌĆō136.



42. Car├®n H, Pollard SM, Beck S. The good, the bad and the ugly: epigenetic mechanisms in glioblastoma. Mol Aspects Med 2013;34:849ŌĆō862.



43. Bastien JI, McNeill KA, Fine HA. Molecular characterizations of glioblastoma, targeted therapy, and clinical results to date. Cancer 2015;121:502ŌĆō516.


44. Zhen L, Yun-Hui L, Hong-Yu D, Jun M, Yi-Long Y. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biol 2016;37:673ŌĆō683.



45. Zhou K, Zhang C, Yao H, Zhang X, Zhou Y, Che Y, et al. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol Cancer 2018;17:105.




46. Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 2006;9:287ŌĆō300.


47. Bredel M, Bredel C, Juric D, Harsh GR, Vogel H, Recht LD, et al. Functional network analysis reveals extended glioma-genesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res 2005;65:8679ŌĆō8689.


48. Ma Y, Zhou G, Li M, Hu D, Zhang L, Liu P, et al. Long noncoding RNA DANCR mediates cisplatin resistance in glioma cells via activating AXL/PI3K/Akt/NF-kappaB signaling pathway. Neurochem Int 2018;118:233ŌĆō241.


49. Li J, Zhou L. Overexpression of lncRNA DANCR positively affects progression of glioma via activating Wnt/beta-catenin signaling. Biomed Pharmacother 2018;102:602ŌĆō607.


50. Wang R, Zhang T, Yang Z, Jiang C, Seng J. Long non-coding RNA FTH1P3 activates paclitaxel resistance in breast cancer through miR-206/ABCB1. J Cell Mol Med 2018;22:4068ŌĆō4075.



51. Feng T, Shao F, Wu Q, Zhang X, Xu D, Qian K, et al. miR-124 downregulation leads to breast cancer progression via LncRNA-MALAT1 regulation and CDK4/E2F1 signal activation. Oncotarget 2016;7:16205ŌĆō16216.



52. Wang Y, Zhou Y, Yang Z, Chen B, Huang W, Liu Y, et al. MiR-204/ZEB2 axis functions as key mediator for MALAT1-induced epithelial-mesenchymal transition in breast cancer. Tumour Biol 2017;39:1010428317690998.



53. Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 2007;449:557ŌĆō563.



54. Zhao H, Langerod A, Ji Y, Nowels KW, Nesland JM, Tibshirani R, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell 2004;15:2523ŌĆō2536.



55. Deng X, Zhao Y, Wu X, Song G. Upregulation of CCAT2 promotes cell proliferation by repressing the P15 in breast cancer. Biomed Pharmacother 2017;91:1160ŌĆō1166.


- TOOLS
-
METRICS

-
- 44 Crossref
- 0 Scopus
- 8,676 View
- 284 Download
- Related articles in GNI