Generation of Whole-Genome Sequencing Data for Comparing Primary and Castration-Resistant Prostate Cancer
Article information
Abstract
Because castration-resistant prostate cancer (CRPC) does not respond to androgen deprivation therapy and has a very poor prognosis, it is critical to identify a prognostic indicator for predicting high-risk patients who will develop CRPC. Here, we report a dataset of whole genomes from four pairs of primary prostate cancer (PC) and CRPC samples. The analysis of the paired PC and CRPC samples in the whole-genome data showed that the average number of somatic mutations per patients was 7,927 in CRPC tissues compared with primary PC tissues (range, 1,691 to 21,705). Our whole-genome sequencing data of primary PC and CRPC may be useful for understanding the genomic changes and molecular mechanisms that occur during the progression from PC to CRPC.
Introduction
Prostate cancer (PC) is the most common malignancy in males [1]. It is known that about 20% of PC patients experience disease progression and distant metastasis [2, 3]. The therapeutic options for patients with aggressive PC include prostatectomy, radiation therapy, and androgen deprivation therapy (ADT) [4]. Although ADT therapy induces short 2–3-year remissions, unfortunately, most PCs eventually progress into castration-resistant prostate cancer (CRPC) [3], which does not respond to ADT therapy and shows poor clinical behavior. Therefore, it is crucial to understand the molecular characteristics and identify robust biomarkers that are associated with the development of CRPC from primary PC.
High-throughput next-generation sequencing technologies have gradually uncovered the molecular characteristics of PC, along with CRPC [5–8]. However, many genomic studies on CRPC have been conducted on metastasized CRPC that has been discovered at distant organs. These studies of metastatic sites do not reflect the precise molecular characteristics of CRPC, because these sites are not the primary PC site and because metastatic sites have completely different microenvironments from the primary site [9]. Here, we generated a dataset of whole genomes from four pairs of primary PC and CRPC samples from the same patient (i.e., a total of eight paired samples from four PC patients). In this report, the genomic status of samples with primary PC and CRPC was explored, and different variants between these two distinct phenotypes were identified by comparing the genomes of primary PC and its paired CRPC.
Methods
Tissues samples
Four pairs of primary PC and CRPC tissues were obtained from Chungbuk National University Hospital (Korea) with informed consent and approval of the Internal Review Board at Chungbuk National University. To obtain a consistent variant profile that was associated with the development of CRPC, primary CRPCs were obtained from homogenous biopsy sites, and none of our PC samples was from distant metastatic sites. Detailed clinical characteristics of the four pairs of primary PC and CRPC tissues are described in Supplementary Table 1.
Whole-genome sequencing library construction and sequencing
Genomic DNA was isolated using the DNeasy Blood and Tissue kit (Qiagen, Carlsbad, CA, USA), and the sequencing library was constructed using the Illumina TruSeq DNA Library Prep Kit (San Diego, CA, USA). Next, paired-end sequencing was performed on an Illumina HiSeq X Ten sequencing instrument, yielding ~ 150-bp short sequencing reads.
Data analysis
The sequenced reads were aligned to human reference genome 19 using Burrows Wheelers Aligner [10], and duplicate reads were removed using Picard (Broad Institute). Then, the remaining reads were calibrated and realigned using the Genome Analysis Toolkit [11]. The realigned Binary Alignment Map files were analyzed using Strelka [12] to detect somatic single-nucleotide variants and insertions/ deletions. For all programs, the default parameter settings were applied.
Results and Discussion
Quality and quantity of the sequencing data
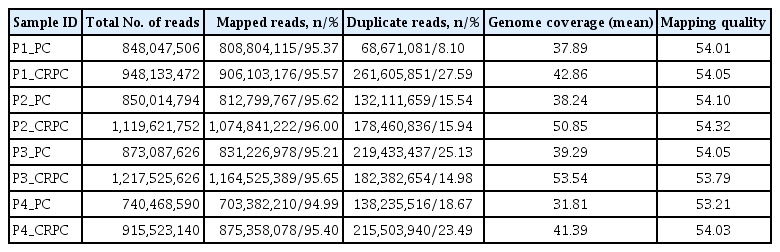
The whole-genome sequencing (WGS) data, including the mapping rate, genome coverage, scores of the mapping quality, and duplicate reads, are summarized in Table 1. Briefly, the mapping rate and scores of the mapping quality in the four pairs of primary PC and CRPC samples were higher than 95% and 53%, respectively. In addition, the average genome coverage of our samples was over 30× (between 31.81× and 53.54×). Although coverage of several hundred times is required for detecting low-level mutations in next-generation sequence data [13], WGS with 30× sequence coverage is appropriate for comprehensive identification of tumor-specific somatic mutations [14]. These results suggest that the quality and quantity of our sequencing data are adequate for mutational analysis during the progression from PC to CRPC.
Mutation patterns identified from CRPC compared with PC
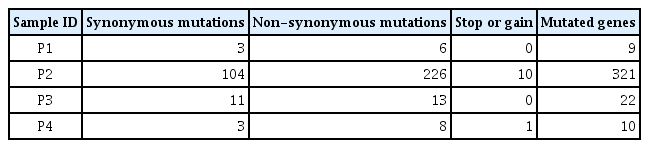
The average number of somatic mutations per patients was 7,927 in CRPC tissues compared with primary PC tissues (range, 1691 to 21,705). In particular, patient P2 had hypermutations (n = 21,705), whereas patient P1 had a low mutation frequency (n = 1,691) (Fig. 1A). To observe the mutation signatures in the development of CRPC from primary PC, we examined the spectrum of base substitutions. This analysis revealed an unusually high proportion of C:G > T:A and A:T>G:C transversions (Fig. 1B), similar to a previous study [15]. Next, the mutated sites were annotated as non-synonymous, synonymous, stop, and gain mutations. The number of mutations affecting protein-coding genes was 9, 321, 22, and 10 for the four patients (Table 2), and we observed recurrent mutations in the ANKRD20A4, ANDRK38B, AQP7, GGT1, and TAS2R31 genes. Detailed information for the non-synonymous and recurrent mutations is summarized in Supplementary Table 2. Further study will be needed to examine whether the mutated genes are associated with the development of CRPC from primary PC.

Number of mutations and distribution of mutation type. (A) Somatic mutations were detected using the Strelka package with default parameter settings. (B) Relative distribution of single-base substitutions by type in each of the four paired castration-resistant prostate cancer patients. SNV, single nucleotide variant.
In conclusion, PC is a heterogeneous disease and has various steps in its disease progression, including CRPC, the poorest prognostic status during the progression of PC. Understanding the molecular characteristics of the development of CRPC will help identify high-risk PC patients and develop novel therapeutic strategies to block the progression of CRPC. We generated a set of WGS data, consisting of eight PC samples containing four pairs of primary PC and CRPC samples from the same patient, because genetic mutations have the greatest potential to play a role in the progression of PC and CRPC and the therapeutic management of CRPC [16, 17]. By comparing primary PC and its paired CRPC, many somatic mutations that were significantly associated with the development of CRPC were identified, including TP53 and KMT2C, which are known to be involved in the progression of PC [16, 17]. We hope that our whole-genome sequence data of the four paired PC and CRPC tissues will be utilized by many researchers to understand the progression of PC and the resistance to androgen deprivation therapy.
Notes
Availability: The whole-genome data are available in the Korean Bioinformation Center (KOBIC) biodata (http://biodata.kr/) public database under accession numbers KBRS20180406_0000001–KBRS20180406_0000008.
Authors’ contribution
Conceptualization: SYK
Sample and data curation: SJY, WJK, WTK, PJ, HWK
WGS data generation: JHK, PJ
Data analysis: JLP, SKK
Writing – original draft: JLP, SKK
Writing – review & editing: SYK
Acknowledgments
This research was supported by a National Research Foundation of Korea (NRF) grant (NRF-2014M3C9A3068 554 and NRF-2017MBA9B5060884), funded by the Korean government (MEST), and a grant from the KRIBB Research Initiative Program.
Supplementary Materials
Supplementary data including two tables can be found with this article online at https://doi.org/10.5808/GI.2018.16.3.71.