|
 |
- Search
| Genomics Inform > Volume 15(4); 2017 > Article |
|
Abstract
Apurinic/apyrimidinic endonuclease 1 (APE1) is an enzyme responsible for the initial step in the base excision repair pathway and is known to be a potential drug target for treating cancers, because its expression is associated with resistance to DNA-damaging anticancer agents. Although several inhibitors already have been identified, the identification of novel kinds of potential inhibitors of APE1 could provide a seed for the development of improved anticancer drugs. For this purpose, we first classified known inhibitors of APE1. According to the classification, we constructed two distinct pharmacophore models. We screened more than 3 million lead-like compounds using the pharmacophores. Hits that fulfilled the features of the pharmacophore models were identified. In addition to the pharmacophore screen, we carried out molecular docking to prioritize hits. Based on these processes, we ultimately identified 1,338 potential inhibitors of APE1 with predicted binding affinities to the enzyme.
DNA damage occurs naturally and due to the environment, altering the cell’s abilities that are encoded by the DNA, and may lead to diseases, like cancer. Cells respond to DNA damage by DNA repair and cellular apoptosis [1, 2]. Apurinic/apyrimidinic endonuclease (APE) is an enzyme that identifies damaged apurinic/apyrimidinic sites in DNA, cuts the phosphodiester bond in the backbone of the sites, and has critical roles in the base excision pathway [3]. APE1 has recently been noted as a potential drug target for treating cancer, in that overexpression of the enzyme has been observed and shown to be associated with a poor response to cancer treatment, such as radiation and anticancer drugs, and a lower overall survival rate [4–7]. Antineoplastic agents that are to treat cancers are known to induce the expression of APE1, increasing the resistance of tumor cells to drug treatment. Thus, compounds that inhibit the activity of APE1 could be potential anticancer drugs with DNA-damaging antineoplastic agents used in the clinic [8].
For this reason, there have been several attempts to develop compounds targeting APE1. Currently, although there is no approved drug yet, three candidates—7-nitroindole-2-carboxylic acid (also known as CRT0044876) [9, 10], lucanthone (also known as Miracil D) [9], and methoxyamine (trademark TRC102)—are known to inhibit APE1 activity and are under examination in clinical trials. Lucanthone and CRT0044876 have rings similar to the deoxyribose sugar ring without a base and many hydrogen bond acceptors that can interact with hydrogen bond donors in the active site of APE1. These properties lead APE1 to stick in the site and prevent it from repairing DNA damage [11]. Methoxyamine is known to attack the open-ring form of AP sites to form an oxime linkage. In other words, methoxyamine blocks APE1 from accessing the lesion site rather than targeting the enzyme directly. This may lead to nonspecific off-target effects [12, 13]. Although several inhibitors of APE1 have been discovered, most potent compounds have weaknesses [14]. Thus, it is necessary to find novel kinds of potential inhibitors targeting APE1. Here, we present out work, in which we applied pharmacophore modeling and virtual screening. The overall procedures we carried out are illustrated in Fig. 1. We constructed pharmacophore models by capturing the common features of known inhibitors of APE1. The modes were used to screen a vast number of lead-like compounds, and molecular docking was used to prioritize the hits of the screen.
From the ChEMBL [15] database, we retrieved 52 compounds known to be targets of APE1 and 51 compounds with an IC50 of less than 10 μM. By eliminating redundancy, the number of compounds was reduced into 83. The list did not contain methoxyamine; so, methoxyamine was also added to the list.
We clustered these 84 compounds by Tanimoto distance, based on the PubChem fingerprint [16], and finally categorized them into two groups by excluding two outliers (CHEMBL1213633 and CHEMBL313493) (Fig. 2). A total of 49 molecules in group 1 (Fig. 3) and 33 molecules in group 2 (Fig. 4) were used to generate pharmacophore models 1 and 2, respectively.
Ligandscout tools 4.1 [17] was used to generate the ligand-based pharmacophore models. Ligandscout is known to be able to increase the selectivity of a pharmacophore model with the excluded volume feature. To generate more flexible pharmacophore, the threshold of the portion of partially matching features was set to 20%.
For the initial set of the pharmacophore screen, we selected a lead-like subset [18], defined by the ZINC database (ZINC is not commercial) [19]. Similar to druglikeness [20], like Lipinski’s rule of 5 [21], lead-like compounds are defined as being large enough to be validated in experiments but are smaller than most drugs, optimized too specifically, and more soluble than their drug-like compounds. ZINC provides a lead-like subset fulfilling leadlikeness as follows: (1) molecular weight between 250 and 350 Da, (2) partition coefficient log p ≤ 3.5, and (3) no more than seven rotatable bonds. The structures of lead-like compounds in medium pH were downloaded and converted into a database for screening by idbgen, a component of Ligandscout. We carried out pharmacophore screens using iscreen in Ligandscout for models 1 and 2 independently. Pharmacophore fit scores were also calculated by LigandScout based on the number of matching pharmacophore features and the root-mean-square deviation of the pharmacophore alignment.
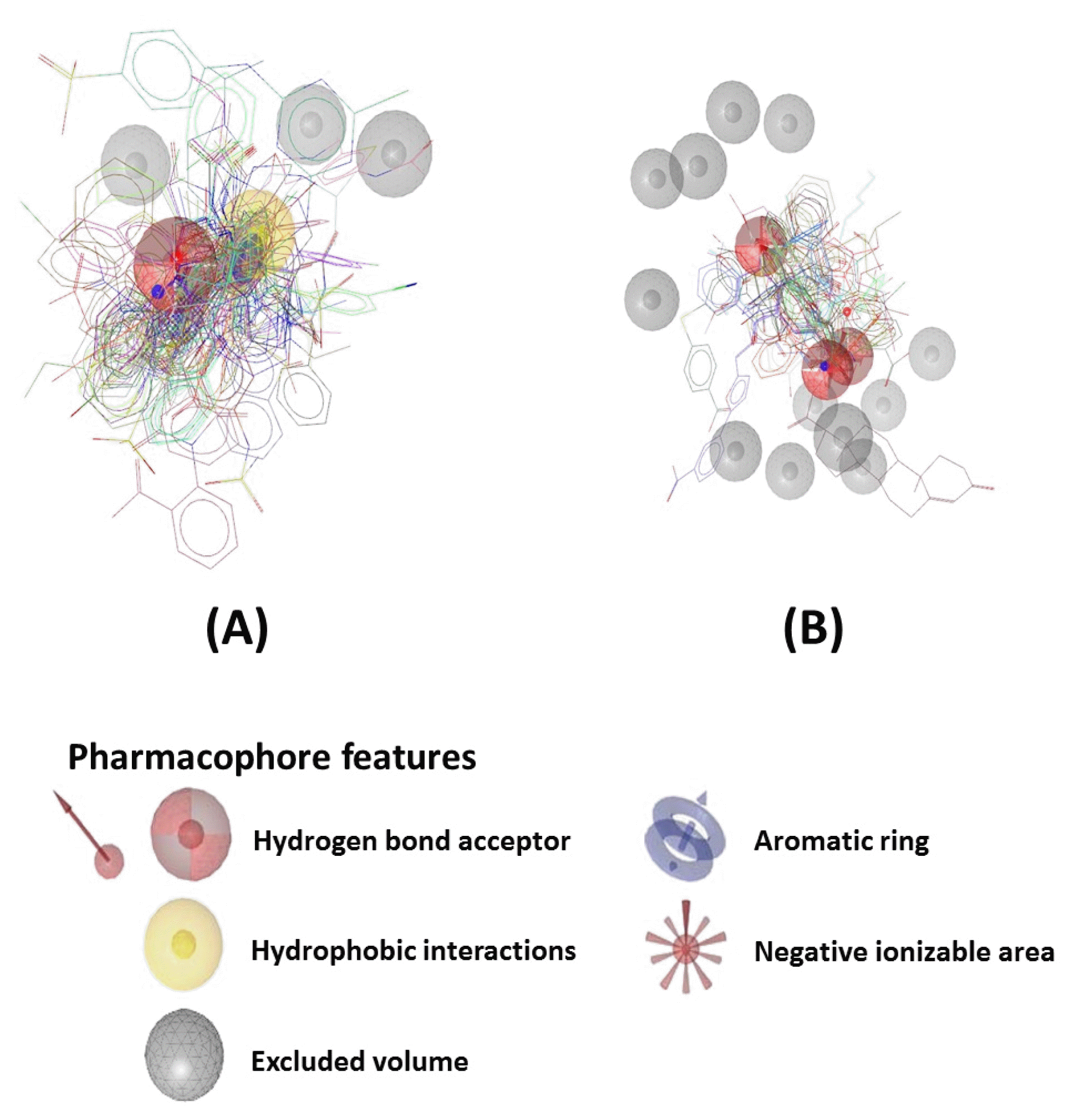
A total of 84 compounds from the ChEMBL database were first collected to generate a pharmacophore, but their structures and properties were too heterogeneous to get common features. Thus, we carried out clustering and categorized the compounds into two groups (Figs. 2,–4). For each group of compounds, we generated the corresponding pharmacophore model. Pharmacophore model 1 was generated by 49 compounds from group 1. The model was composed of four features (one hydrophobic centroid, one aromatic ring, two hydrogen acceptors) and three exclusion volume spaces (Fig. 5A). Model 2 was generated by 33 compounds from group 2. The model was composed of four features (one negative ionizable and three hydrogen bond acceptors) and 12 exclusion volume spaces (Fig. 5B).
For 3,563,829 lead-like compounds retrieved from the ZINC database, we performed a pharmacophore screen based on pharmacophore models 1 and 2 independently. Among multiple subsets provided by ZINC, we chose the lead-like subset, not the drug-like set, because we aimed to provide a list of potential hits that could be optimized further by other groups, as well as our group.
As a result, 400,153 and 290,742 hits fulfilled the features of models 1 and 2, respectively. The intersection of the two lists of hits, which fulfilled all features of both models, consisted of 38,087 compounds. To remove structurally similar compounds, we clustered the 38,087 hits by hierarchical clustering, based on the Tanimoto distance in PubChem Fingerprint. According to the result of the clustering, we ruled out redundant compounds that had similar compounds (Tanimoto coefficient >0.8). Thus, 1,338 hits eventually remained as potential inhibitors of APE1.
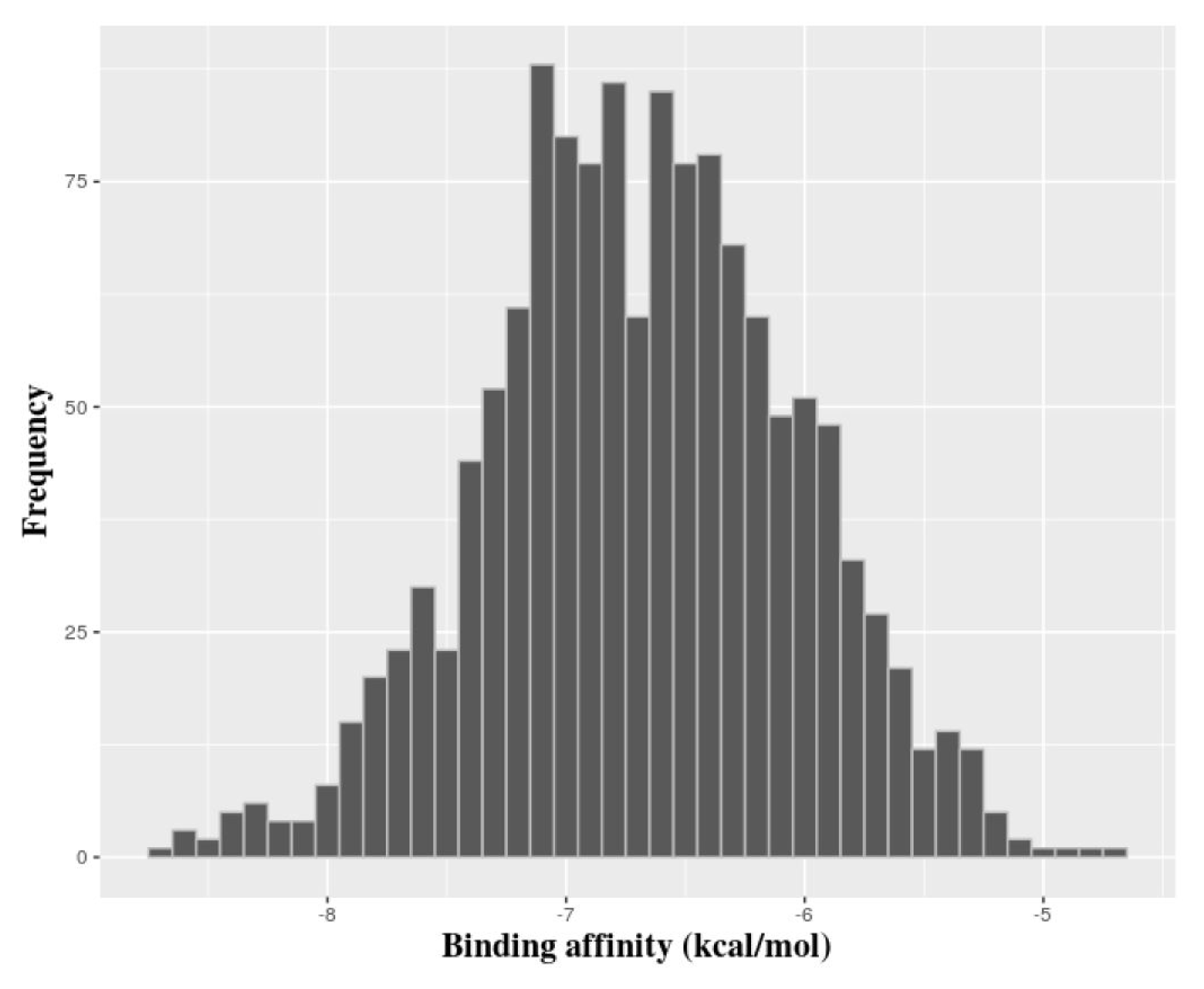
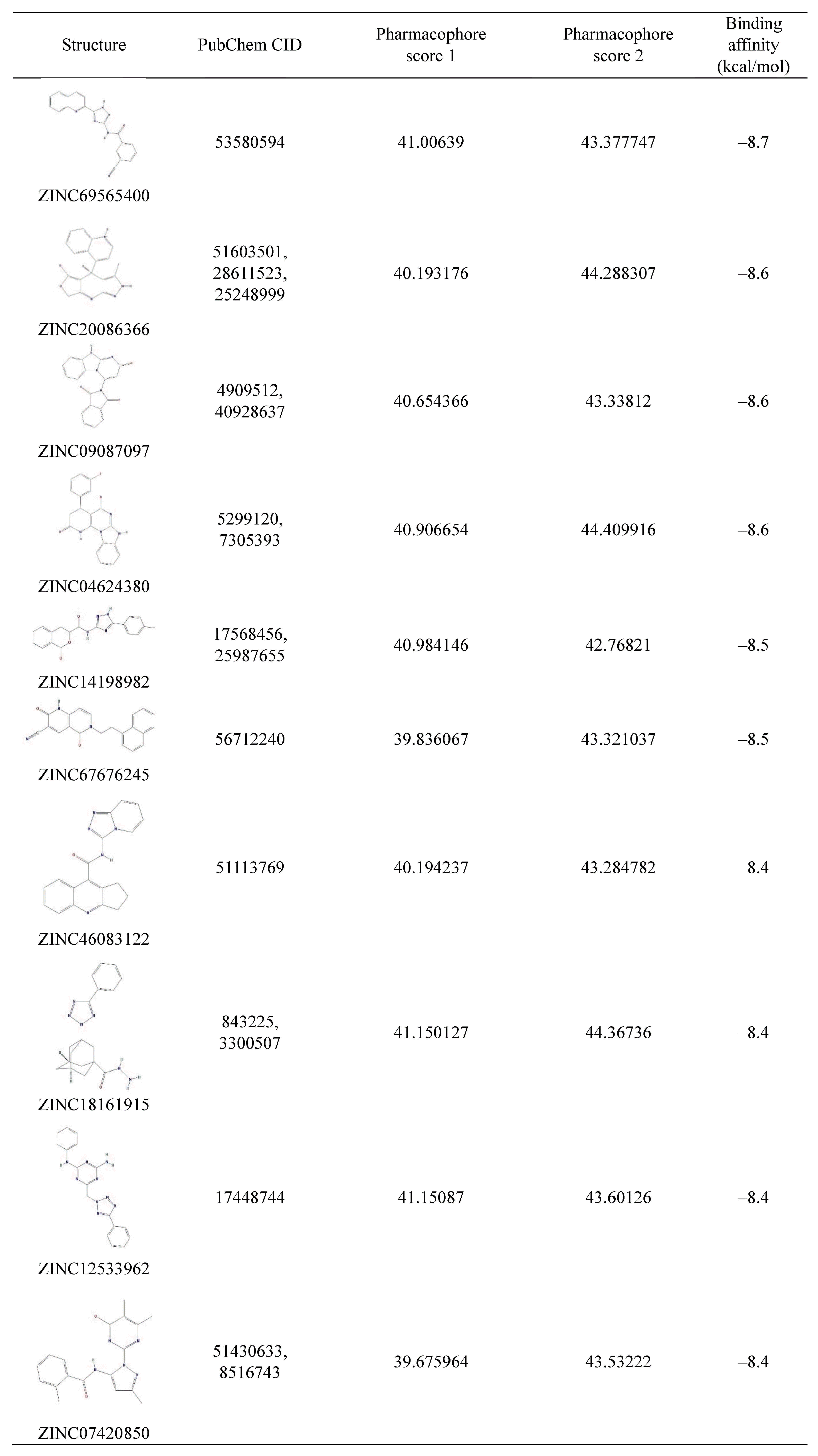
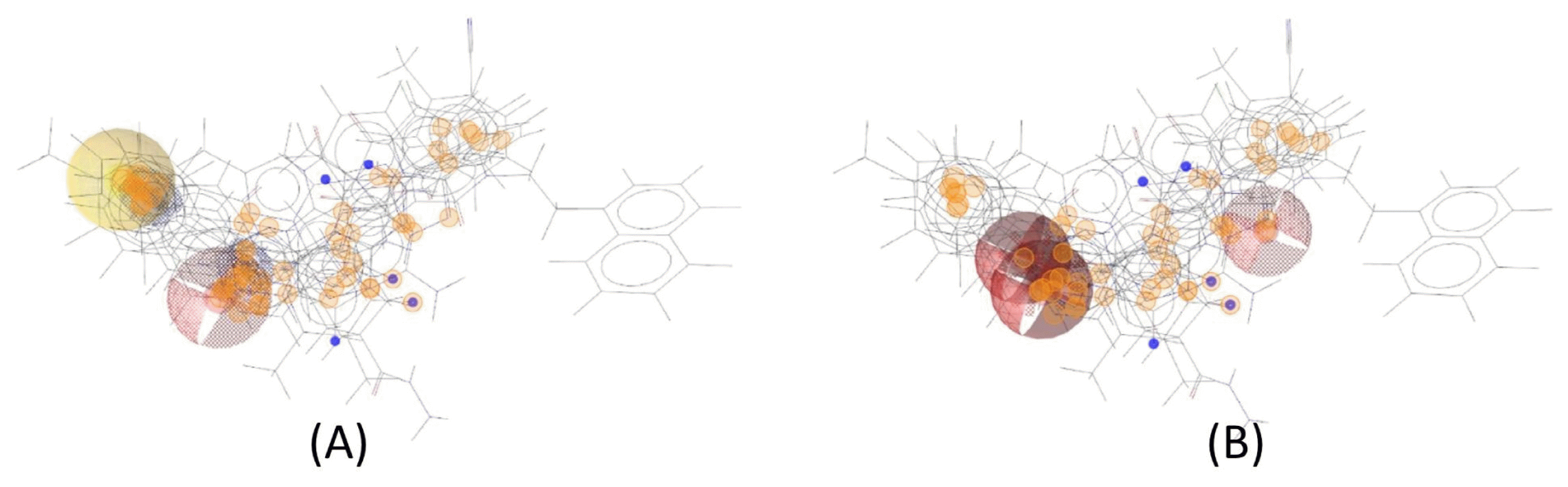
We carried out molecular docking of the hits against APE1 to prioritize the hits using AutoDock Vina. Fig. 6 depicts the distribution of the predicted binding energies of the hits of the pharmacophore screen by docking. After molecular docking, we did not filter out compounds based on a particular threshold of the predicted value of the binding affinity but instead provide the top 10 hits in Fig. 7, their predicted binding poses in Supplementary Fig. 1, and all of the hits in Supplementary Table 1. This is because although Shityakov and Förster [25] reported that a compound having a binding affinity predicted by AutoDock Vina of lower than −6 kcal/mol could be considered an active hit, the values are only predictive and rely on a somewhat empirical energy function. In other words, predicted binding affinities should be used restrictedly to help those who want to validate hits to determine the priority of subjects of an assay. Fig. 8 shows the alignments of the best hits into each pharmacophore model; all of the hits map well to the pharmacophore models. Of note, the rank of the docking results does not mean pharmacophore fitness, and all of the inhibitor compounds we found here can be mapped to the models well. The figure of pharmacophore alignment was made to provide an example showing that our hits can be mapped properly.
In summary, we screened more than 3 million lead-like compounds by pharmacophore modeling, and 1,338 hits were suggested to be potential inhibitors of APE1. However, this work has a limitation, due to the lack of experimental validation. Nevertheless, the list of hits in this work could reduce the time and cost of researchers who want to develop novel anticancer drugs inhibiting the activity of APE1, since we prioritized candidates of the experiments and since all of them have lead-like properties, which means that the hits are appropriate for further optimization and development into drugs.
Currently, there are several approaches that apply hits from a pharmacophore screen for further development in to a novel drug. Fei et al. [26] first developed a pharmacophore model of a drug target, like our method; then, 3D-quantitative structure-activity relationship (QSAR) modeling was used for validation and further virtual screening. Wieder et al. [27] proposed a novel approach combining pharmacophore modeling and molecular dynamics (MD) simulations, and they showed that their methods were likely to result in more robust hits. Like these approaches, the results from pharmacophore modeling could be adopted in other in silico methods, such as molecular docking, QSAR modeling, and MD simulation. It is worth combining these methods and our results to get more robust results. If further integrative approaches and in vitro or vivo assays of hits validate our results, our method could be applied to other drug targets, in addition to APE1.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF), funded by the Ministry of Science and ICT (NRF-2017R1C1B2008617 and NRF-2017M3A9B6061511), KREONET (Korea Research Environment Open NETwork) which is managed and operated by KISTI (Korea Institute of Science and Technology Information). In Won Lee was supported by a Sangji University scholarship for research assistants.
Supplementary materials
Supplementary data including one table and one figure can be found with this article online at http://www.genominfo.org/src/sm/gni-15-147-s001.pdf.
Supplementary Fig. 1.
Predicted binding poses of the top 10 hits and AP endonuclease 1.
Supplementary Table 1.
List of 1,338 potential inhibitors of AP endonuclease 1 by pharmacophore screening and molecular docking
Fig. 1
Outline of overall procedures of the screen to find potential inhibitors of apurinic/apyrimidinic endonuclease 1.

Fig. 5
Pharmacophore models used for screening. Models were generated by compounds in group 1 (A) and group 2 (B).
References
1. Bjorksten J, Acharya PV, Ashman S, Wetlaufer DB. Gerogenic fractions in the tritiated rat. J Am Geriatr Soc 1971;19:561–574.


2. Acharya PV. The isolation and partial characterization of age-correlated oligo-deoxyribo-ribonucleotides with covalently linked aspartyl-glutamyl polypeptides. Johns Hopkins Med J Suppl 1972;(1):254–260.

3. Barzilay G, Hickson ID. Structure and function of apurinic/apyrimidinic endonucleases. Bioessays 1995;17:713–719.


4. Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci U S A 1996;93:8919–8923.



5. Ludwig DL, MacInnes MA, Takiguchi Y, Purtymun PE, Henrie M, Flannery M, et al. A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat Res 1998;409:17–29.


6. Kelley MR, Logsdon D, Fishel ML. Targeting DNA repair pathways for cancer treatment: what’s new? Future Oncol 2014;10:1215–1237.



7. Seiple LA, Cardellina JH 2nd, Akee R, Stivers JT. Potent inhibition of human apurinic/apyrimidinic endonuclease 1 by arylstibonic acids. Mol Pharmacol 2008;73:669–677.


8. Srinivasan A, Wang L, Cline CJ, Xie Z, Sobol RW, Xie XQ, et al. Identification and characterization of human apurinic/apyrimidinic endonuclease-1 inhibitors. Biochemistry 2012;51:6246–6259.



9. Naidu MD, Agarwal R, Pena LA, Cunha L, Mezei M, Shen M, et al. Lucanthone and its derivative hycanthone inhibit apurinic endonuclease-1 (APE1) by direct protein binding. PLoS One 2011;6:e23679.



10. Simeonov A, Kulkarni A, Dorjsuren D, Jadhav A, Shen M, McNeill DR, et al. Identification and characterization of inhibitors of human apurinic/apyrimidinic endonuclease APE1. PLoS One 2009;4:e5740.



11. Luo M, Kelley MR. Inhibition of the human apurinic/apyrimidinic endonuclease (APE1) repair activity and sensitization of breast cancer cells to DNA alkylating agents with lucanthone. Anticancer Res 2004;24:2127–2134.

12. Liuzzi M, Talpaert-Borlé M. A new approach to the study of the base-excision repair pathway using methoxyamine. J Biol Chem 1985;260:5252–5258.


13. Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol Aspects Med 2007;28:375–395.


14. Wilson DM 3rd, Simeonov A. Small molecule inhibitors of DNA repair nuclease activities of APE1. Cell Mol Life Sci 2010;67:3621–3631.



15. Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 2012;40:D1100–D1107.


16. Wang Y, Bryant SH, Cheng T, Wang J, Gindulyte A, Shoemaker BA, et al. PubChem BioAssay: 2017 update. Nucleic Acids Res 2017;45:D955–D963.



17. Wolber G, Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model 2005;45:160–169.


18. Teague SJ, Davis AM, Leeson PD, Oprea T. The design of lead-like combinatorial libraries. Angew Chem Int Ed Engl 1999;38:3743–3748.


19. Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model 2012;52:1757–1768.



21. Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 2001;46:3–26.


22. Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature 2000;403:451–456.



23. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455–461.



24. Le Guilloux V, Schmidtke P, Tuffery P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics 2009;10:168.



25. Shityakov S, Förster C.
In silico predictive model to determine vector-mediated transport properties for the blood-brain barrier choline transporter. Adv Appl Bioinform Chem 2014;7:23–36.


- TOOLS
-
METRICS

-
- 1 Crossref
- 0 Scopus
- 8,432 View
- 198 Download