 |
 |
- Search
| Genomics Inform > Volume 10(2); 2012 > Article |
Abstract
Acknowledgments
Supplementary materials
References
Fig.┬Ā1

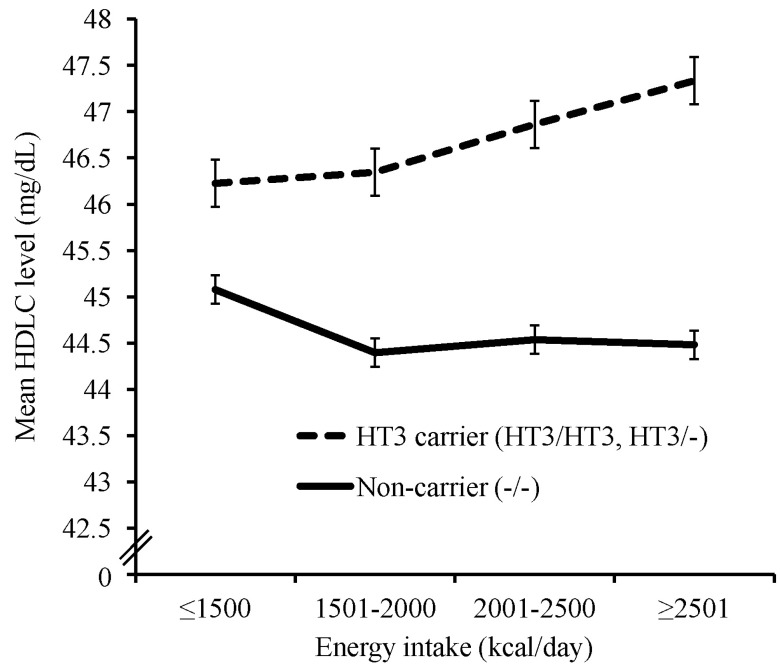
Fig.┬Ā2

Fig.┬Ā3
Table┬Ā1.
| Parameter |
Stage 1 |
Stage 2 |
||
|---|---|---|---|---|
| Ansung (n = 3,434) | Ansan (n = 4,102) | Total (n = 7,536) | Total (n = 3,703) | |
| Agea | 54.8 ┬▒ 8.8 | 48.2 ┬▒ 7.4 | 51.2 ┬▒ 8.7 | 53.2 ┬▒ 8.3 |
| Gender (%) | ||||
| ŌĆāMalea | 1,534 (44.7) | 2,104 (51.3) | 3,638 (48.3) | 1,651 (44.6) |
| ŌĆāFemalea | 1,900 (55.3) | 1,998 (48.7) | 3,898 (51.7) | 2,052 (55.4) |
| BMI (kg/m2)a | 24.2 ┬▒ 3.2 | 24.5 ┬▒ 2.9 | 24.4 ┬▒ 3.1 | 24.0 ┬▒ 2.9 |
| Systolic blood pressure (mm Hg)a | 123.8 ┬▒ 17.6 | 115.4 ┬▒ 16.3 | 119.2 ┬▒ 17.4 | 121.9 ┬▒ 14.8 |
| Diastolic blood pressure (mm Hg)a | 81.3 ┬▒ 10.6 | 77.4 ┬▒ 11.3 | 79.2 ┬▒ 11.2 | 77.2 ┬▒ 10.2 |
| Lipid concentration (mg/dL) | ||||
| ŌĆāHDLCa | 45.1 ┬▒ 10.3 | 45.0 ┬▒ 9.9 | 45.0 ┬▒ 10.1 | 54.6 ┬▒ 13.3 |
| ŌĆāLDLCb | 109.0 ┬▒ 31.0 | 120.6 ┬▒ 31.4 | 115.3 ┬▒ 31.8 | 119.0 ┬▒ 31.4 |
| ŌĆāTGa | 158.8 ┬▒ 103.3 | 156.8 ┬▒ 101.7 | 157.7 ┬▒ 102.4 | 123.3 ┬▒ 91.4 |
| ŌĆāTCHLa | 184.5 ┬▒ 34.5 | 195.7 ┬▒ 35.5 | 190.6 ┬▒ 35.5 | 197.4 ┬▒ 35.1 |
| Energy intake (kcal/day)c | 1,977.6 ┬▒ 916.7 | 1,895.9 ┬▒ 516.0 | 1,932.2 ┬▒ 723.2 | - |
| Physical activity (MET/h)d | 3.8 ┬▒ 1.3 | 2.4 ┬▒ 0.6 | 3.0 ┬▒ 0.7 | - |
| Alcohol consumption (%)e | ||||
| ŌĆāNon-drinker | 1,628 (48.2) | 1,733 (42.3) | 3,361 (45.0) | - |
| ŌĆāEx-drinker | 255 (7.5) | 196 (4.8) | 451 (6.0) | - |
| ŌĆāCurrent drinker | 1,495 (44.3) | 2,164 (52.9) | 3,659 (49.0) | - |
| Smoking status (%)f | ||||
| ŌĆāNon-smoker | 1,987 (59.3) | 2,331 (57.1) | 4,318 (58.1) | - |
| ŌĆāEx-smoker | 403 (12.0) | 724 (17.7) | 1,127 (15.2) | - |
| ŌĆāCurrent smoker | 962 (28.7) | 1,027 (25.2) | 1,989 (26.8) | - |
Quantitative parameters are shown as mean ┬▒ SD or as the number observed, followed by its corresponding percentage.
BMI, body mass index; HDLC, high-density lipoprotein cholesterol; LDLC, low-density lipoprotein cholesterol; TG, triglyceride; TCHL, total cholesterol; MET, metabolic equivalent.
b Missing rates of the parameter are 0.027, 0.025, 0.026, and 0.015 in Ansung, Ansan, total of stage I, and total of stage II, respectively;
c Missing rates of the parameter are 0.056, 0.012, and 0.032 in Ansung, Ansan, and total of stage I, respectively;
d Missing rates of the parameter are 0.021, 0.004, and 0.012 in Ansung, Ansan, and total of stage I, respectively;
Table┬Ā2.
| SNP | Position | Alleles |
Ansung |
Ansan |
Stage 1 (Ansung ’╝ŗ Ansan) |
Stage 2 |
Combined (stage 1 ’╝ŗ 2) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effecta | p-valueb | Effecta | p-valueb | Effecta | p-valuec | Effecta | p-valueb | Effecta | p-valueb | |||
| HDLC-associated SNPs | ||||||||||||
| ŌĆārs263d | 19812812 | G’╝×A | 1.25 | 1.70 ├Ś 10-5 | 1.23 | 1.20 ├Ś 10-6 | 1.23 | 1.20 ├Ś 10-10 | 1.53 | 5.50 ├Ś 10-6 | 1.22 | 1.70 ├Ś 10-11 |
| ŌĆārs271d | 19813702 | C’╝×T | 1.32 | 5.30 ├Ś 10-6 | 1.24 | 8.30 ├Ś 10-7 | 1.27 | 2.50 ├Ś 10-11 | 1.53 | 5.10 ├Ś 10-6 | 1.24 | 5.70 ├Ś 10-12 |
| ŌĆārs17482753e | 19832646 | G’╝×T | 2.27 | 1.90 ├Ś 10-10 | 1.57 | 4.60 ├Ś 10-7 | 1.88 | 1.10 ├Ś 10-15 | 3.21 | 1.20 ├Ś 10-13 | 2.19 | 1.20 ├Ś 10-21 |
| ŌĆārs10503669 | 19847690 | G’╝×T | 2.31 | 1.10 ├Ś 10-10 | 1.53 | 1.30 ├Ś 10-6 | 1.89 | 2.00 ├Ś 10-15 | 3.18 | 1.90 ├Ś 10-13 | 2.24 | 3.60 ├Ś 10-22 |
| ŌĆārs17410962 | 19848080 | G’╝×A | 2.29 | 1.30 ├Ś 10-10 | 1.56 | 5.50 ├Ś 10-7 | 1.88 | 1.00 ├Ś 10-15 | 3.21 | 1.20 ├Ś 10-13 | 2.18 | 1.70 ├Ś 10-21 |
| ŌĆārs17411031 | 19852310 | C’╝×G | 1.59 | 3.80 ├Ś 10-8 | 1.28 | 8.80 ├Ś 10-7 | 1.42 | 2.10 ├Ś 10-13 | 1.94 | 9.40 ├Ś 10-9 | 1.47 | 1.30 ├Ś 10-15 |
| ŌĆārs17489282 | 19852518 | G’╝×A | 1.6 | 2.50 ├Ś 10-8 | 1.19 | 4.70 ├Ś 10-6 | 1.38 | 8.80 ├Ś 10-13 | 1.93 | 1.10 ├Ś 10-8 | 1.45 | 2.70 ├Ś 10-15 |
| ŌĆārs4922117 | 19852586 | A’╝×G | 1.58 | 4.70 ├Ś 10-8 | 1.28 | 8.60 ├Ś 10-7 | 1.42 | 2.50 ├Ś 10-13 | 1.96 | 7.20 ├Ś 10-9 | 1.5 | 3.90 ├Ś 10-16 |
| ŌĆārs17411126 | 19855272 | T’╝×C | 1.57 | 5.60 ├Ś 10-8 | 1.28 | 8.50 ├Ś 10-7 | 1.41 | 2.90 ├Ś 10-13 | 2 | 4.00 ├Ś 10-9 | 1.47 | 1.20 ├Ś 10-15 |
| ŌĆārs765547 | 19866274 | C’╝×T | 1.57 | 5.50 ├Ś 10-8 | 1.27 | 9.60 ├Ś 10-7 | 1.41 | 3.20 ├Ś 10-13 | 1.92 | 1.40 ├Ś 10-8 | 1.46 | 1.40 ├Ś 10-15 |
| ŌĆārs11986942 | 19867445 | G’╝×C | 1.57 | 6.30 ├Ś 10-8 | 1.24 | 1.60 ├Ś 10-6 | 1.39 | 6.20 ├Ś 10-13 | 1.94 | 9.30 ├Ś 10-9 | 1.47 | 1.40 ├Ś 10-15 |
| ŌĆārs1837842 | 19868290 | T’╝×C | 1.57 | 5.50 ├Ś 10-8 | 1.24 | 1.70 ├Ś 10-6 | 1.39 | 5.40 ├Ś 10-13 | 1.92 | 1.40 ├Ś 10-8 | 1.48 | 9.60 ├Ś 10-16 |
| ŌĆārs1919484 | 19869676 | C’╝×T | 1.58 | 5.20 ├Ś 10-8 | 1.23 | 2.20 ├Ś 10-6 | 1.39 | 6.90 ├Ś 10-13 | 1.92 | 1.30 ├Ś 10-8 | 1.46 | 1.90 ├Ś 10-15 |
| ŌĆārs7461115 | 19871540 | G’╝×C | 1.57 | 4.90 ├Ś 10-8 | 1.23 | 2.40 ├Ś 10-6 | 1.39 | 7.20 ├Ś 10-13 | 2.01 | 4.00 ├Ś 10-9 | 1.5 | 4.80 ├Ś 10-16 |
| ŌĆārs7013777 | 19878356 | A’╝×G | 1.51 | 1.40 ├Ś 10-7 | 1.28 | 6.80 ├Ś 10-7 | 1.39 | 4.80 ├Ś 10-13 | 2.05 | 1.80 ├Ś 10-9 | 1.51 | 2.50 ├Ś 10-16 |
| TG-associated SNPs | ||||||||||||
| ŌĆārs263d | 19812812 | G’╝×A | ŌłÆ8.73 | 5.10 ├Ś 10-4 | ŌłÆ8.82 | 9.30 ├Ś 10-4 | ŌłÆ8.79 | 1.70 ├Ś 10-6 | ŌłÆ8.58 | 3.30 ├Ś 10-3 | ŌłÆ8.13 | 6.80 ├Ś 10-7 |
| ŌĆārs271d | 19813702 | C’╝×T | ŌłÆ8.81 | 3.90 ├Ś 10-4 | ŌłÆ9.14 | 5.20 ├Ś 10-4 | ŌłÆ9 | 7.20 ├Ś 10-7 | ŌłÆ8.71 | 2.20 ├Ś 10-3 | ŌłÆ8.31 | 2.70 ├Ś 10-7 |
| ŌĆārs17482753e | 19832646 | G’╝×T | ŌłÆ13.56 | 1.60 ├Ś 10-6 | ŌłÆ16.01 | 6.60 ├Ś 10-8 | ŌłÆ14.83 | 6.00 ├Ś 10-13 | ŌłÆ12.97 | 3.30 ├Ś 10-6 | ŌłÆ13.59 | 4.60 ├Ś 10-15 |
| ŌĆārs10503669 | 19847690 | G’╝×T | ŌłÆ14.04 | 7.20 ├Ś 10-7 | ŌłÆ15.79 | 1.10 ├Ś 10-7 | ŌłÆ14.94 | 4.60 ├Ś 10-13 | ŌłÆ12.44 | 8.20 ├Ś 10-6 | ŌłÆ13.64 | 3.00 ├Ś 10-15 |
| ŌĆārs17410962 | 19848080 | G’╝×A | ŌłÆ13.58 | 1.60 ├Ś 10-6 | ŌłÆ16.3 | 2.90 ├Ś 10-8 | ŌłÆ14.99 | 2.80 ├Ś 10-13 | ŌłÆ12.52 | 6.90 ├Ś 10-6 | ŌłÆ13.53 | 6.40 ├Ś 10-15 |
| ŌĆārs17411031 | 19852310 | C’╝×G | ŌłÆ10.26 | 1.90 ├Ś 10-5 | ŌłÆ10.19 | 6.20 ├Ś 10-5 | ŌłÆ10.26 | 4.50 ├Ś 10-9 | ŌłÆ12.18 | 6.80 ├Ś 10-7 | ŌłÆ10.27 | 3.70 ├Ś 10-12 |
| ŌĆārs17489282 | 19852518 | G’╝×A | ŌłÆ10.42 | 1.80 ├Ś 10-5 | ŌłÆ9.75 | 1.30 ├Ś 10-4 | ŌłÆ10.09 | 9.60 ├Ś 10-9 | ŌłÆ11.92 | 1.40 ├Ś 10-6 | ŌłÆ10.13 | 9.40 ├Ś 10-12 |
| ŌĆārs4922117 | 19852586 | A’╝×G | ŌłÆ10.25 | 1.70 ├Ś 10-5 | ŌłÆ9.94 | 1.00 ├Ś 10-4 | ŌłÆ10.12 | 6.80 ├Ś 10-9 | ŌłÆ12.22 | 6.80 ├Ś 10-7 | ŌłÆ10.28 | 2.80 ├Ś 10-12 |
| ŌĆārs17411126 | 19855272 | T’╝×C | ŌłÆ10.29 | 1.80 ├Ś 10-5 | ŌłÆ10.17 | 7.30 ├Ś 10-5 | ŌłÆ10.27 | 5.10 ├Ś 10-9 | ŌłÆ12.59 | 3.10 ├Ś 10-7 | ŌłÆ10.36 | 2.90 ├Ś 10-12 |
| ŌĆārs765547 | 19866274 | C’╝×T | ŌłÆ10.31 | 1.70 ├Ś 10-5 | ŌłÆ9.6 | 6.20 ├Ś 10-5 | ŌłÆ9.98 | 4.10 ├Ś 10-9 | ŌłÆ11.9 | 2.10 ├Ś 10-6 | ŌłÆ10.01 | 6.70 ├Ś 10-12 |
| ŌĆārs11986942 | 19867445 | G’╝×C | ŌłÆ10.28 | 1.90 ├Ś 10-5 | ŌłÆ9.43 | 9.40 ├Ś 10-5 | ŌłÆ9.87 | 6.80 ├Ś 10-9 | ŌłÆ11.95 | 1.90 ├Ś 10-6 | ŌłÆ9.99 | 7.70 ├Ś 10-12 |
| ŌĆārs1837842 | 19868290 | T’╝×C | ŌłÆ10.23 | 2.00 ├Ś 10-5 | ŌłÆ9.18 | 1.10 ├Ś 10-4 | ŌłÆ9.72 | 8.10 ├Ś 10-9 | ŌłÆ12.12 | 1.60 ├Ś 10-6 | ŌłÆ9.98 | 5.50 ├Ś 10-12 |
| ŌĆārs1919484 | 19869676 | C’╝×T | ŌłÆ10.36 | 1.60 ├Ś 10-5 | ŌłÆ9.34 | 1.00 ├Ś 10-4 | ŌłÆ9.86 | 6.30 ├Ś 10-9 | ŌłÆ11.96 | 1.80 ├Ś 10-6 | ŌłÆ9.98 | 7.10 ├Ś 10-12 |
| ŌĆārs7461115 | 19871540 | G’╝×C | ŌłÆ10.24 | 3.10 ├Ś 10-5 | ŌłÆ9.14 | 1.20 ├Ś 10-4 | ŌłÆ9.71 | 1.40 ├Ś 10-8 | ŌłÆ11.7 | 3.70 ├Ś 10-6 | ŌłÆ9.83 | 2.10 ├Ś 10-11 |
| ŌĆārs7013777 | 19878356 | A’╝×G | ŌłÆ10.37 | 2.90 ├Ś 10-5 | ŌłÆ9.56 | 7.40 ├Ś 10-5 | ŌłÆ9.99 | 7.50 ├Ś 10-9 | ŌłÆ12.02 | 1.20 ├Ś 10-6 | ŌłÆ10.12 | 5.20 ├Ś 10-12 |
SNP, single-nucleotide polymorphism; HDLC, high-density lipoprotein cholesterol; TG, triglyceride; LPL, lipoprotein lipase; LD, linkage disequilibrium.
a Unstandardized coefficients of untransformed values (effect). Effect sizes (mg/dL) of minor alleles assessed in an additive manner;
Table┬Ā3.
| Block | HT |
HT frequency |
HDLC |
TG |
LDLC |
TCHL |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ’╝ŗ’╝ŗ | ’╝ŗ’╝Ź | ’╝Ź’╝Ź | Effecta | p-valueb | Effecta | p-valueb | Effecta | p-valueb | Effecta | p-valueb | ||
| B1 | HT1: AT | 490 (4.4) | 3,645 (32.4) | 7,103 (63.2) | 1.24 | 8.30 ├Ś 10ŌłÆ12 | ŌłÆ8.27 | 3.70 ├Ś 10ŌłÆ7 | 0.92 | 0.08 | 0.96 | 0.11 |
| HT2: GC | 7,065 (62.9) | 3,678 (32.7) | 495 (4.4) | ŌłÆ1.23 | 1.10 ├Ś 10ŌłÆ11 | 8.18 | 5.00 ├Ś 10ŌłÆ7 | ŌłÆ0.96 | 0.06 | ŌłÆ1.03 | 0.09 | |
| B2 | HT1c | 4,888 (43.5) | 5,064 (45.1) | 1,287 (11.5) | ŌłÆ1.19 | 1.05 ├Ś 10ŌłÆ14 | 8.6 | 3.05 ├Ś 10ŌłÆ12 | ŌłÆ0.66 | 0.14 | ŌłÆ0.37 | 0.34 |
| HT2d | 155 (1.4) | 2,427 (21.6) | 8,657 (77.0) | 0.07 | 0.5 | ŌłÆ2.59 | 0.21 | 0.47 | 0.47 | 0.02 | 0.87 | |
| HT3e | 166 (1.5) | 2,295 (20.4) | 8,778 (78.1) | 2.25 | 2.86 ├Ś 10ŌłÆ22 | ŌłÆ13.7 | 9.09 ├Ś 10ŌłÆ15 | 1.21 | 0.06 | 1.1 | 0.08 | |
| HT4f | 71 (0.6) | 1,670 (14.9) | 9,498 (84.5) | 0.08 | 0.62 | ŌłÆ2.85 | 0.39 | ŌłÆ0.68 | 0.38 | ŌłÆ0.83 | 0.33 | |
LPL, lipoprotein lipase; SNP, single-nucleotide polymorphism; HT, haplotype; HDLC, high-density lipoprotein cholesterol; TG, triglyceride; LDLC, low-density lipoprotein cholesterol; TCHL, total cholesterol; B, block.
a Unstandardized coefficients of untransformed values (effect). Effect sizes (mg/dL) of haplotypes assessed in an additive manner;
- TOOLS
-
METRICS

-
- 9 Crossref
- 6,031 View
- 63 Download
- Related articles in GNI





