Web-Based Database and Viewer of East Asian Copy Number Variations
Article information
Abstract
We have discovered copy number variations (CNVs) in 3,578 Korean individuals with the Affymetrix Genome-Wide SNP array 5.0, and 4,003 copy number variation regions (CNVRs) were defined in a previous study. To explore the details of the variants easily in related studies, we built a database, cataloging the CNVs and related information. This system helps researchers browsing these variants with gene and structure variant annotations. Users can easily find specific regions with search options and verify them from system-integrated genome browsers with annotations.
Introduction
Copy number variation (CNV) is a common type of structural variation in the human genome. They have been suggested to be related to disease susceptibility or human phenotype diversity [1-3]. Current genome-wide association studies of CNVs are attempting to find how they are related to disease susceptibility or phenotypic diversity. Due to the fact that frequencies of CNVs show ethnic differences [4-7], a finding of disease or phenotype association of a specific CNV in one population is hard to generalize to other populations. However, by comparing the frequencies of target CNVs among different ethnic groups, we could assume the population-specific disease susceptibility or phenotype difference.
In this regard, CNV frequency information of various populations has become a major concern of population or disease association studies.
With increasing interest in CNVs, many CNV projects have been announced recently. Many of them have already established large databases of CNVs from many different ethnic groups (Database of Genomic Variants [DGV], http://projects.tcag.ca; The Copy Number Variants Projects, http://www.sanger.ac.uk/research/areas/humangenetics/cnv/) [8]. There are also several databases for supporting population-specific studies, including the CNV Control Database for Japanese (http://gwas.lifesciencedb.jp), the Singapore Human Mutation and Polymorphism Database (shmed.bii.a-star.edu.sg) [9], and the Thailand Mutation and Variation Database (www4a.biotec.or.th) [10]. The accumulation of CNV information seems promising, in that we can gradually extend the knowledge of CNVs everywhere in the world. However, it will take a long time to fill the gap of CNV databases covering the whole population.
As a step to fill this gap, we discovered a Korean CNV with Affymetrix Genome-Wide SNP array 5.0 (Affymetrix, Santa Clara, CA, USA) in a previous study [11]. In this study, we built a Korean database based on the findings of 4,003 copy number variation regions (CNVRs). To build a tool that is easily accessible via the web for Korean-specific CNV studies, we built a viewer for browsing these CNVs.
Features and Results
System requisites
Java Runtime Environment of Sun Microsystems 1.6.0 (Oracle, Redwood City, CA, USA) or equivalent is required, since the system is written in Java language. We used MySQL database for storing and retrieving CNV data and GBrowse2 [12] for drawing regional information. Both are freely available from their distribution websites.
Since the main search pages are written in Java Server Page (JSP) language, Apache Tomcat is needed for the application server and Apache HTTP server is needed for Gbrowse2 viewer pages (Fig. 1).
Data
CNVs of our previous study were retrieved from the Affymetrix Genome-Wide Human SNP array 5.0 of 3,578 Korean individuals. A total of 4,003 CNVRs were defined, and 2,077 CNVRs (51.9%) were potentially novel. The annotation data for genes were collected based on the Human Mar. 2006 NCBI36/hg18 build, and reference structure variants were retrieved from DGV (hg18.v8.aut.2009).
Database and viewer
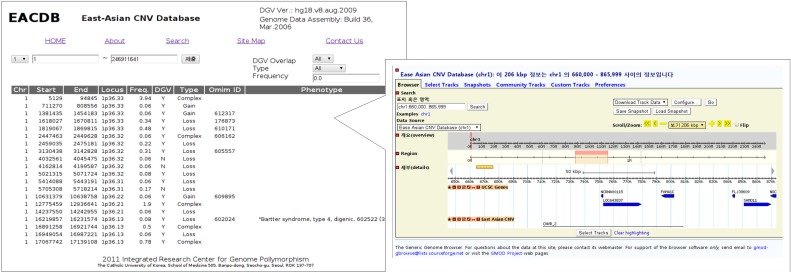
Our web-based database viewer can display previously discovered CNVs by their positions (Fig. 2). Users can also filter out CNVs based on the DGV overlapped regions, CNV type (Gain/Loss/Complex), or their frequencies. Each selected region could be diagnosed in detail by clicking on it. DGV and OMIM ID columns are linked with corresponding websites, and CNVR position columns are linked with the genome browser. The genome browser is integrated based on the open source project GBrowse2. Users can seek or zoom in/out of CNVs across the chromosome by entering positions or clicking zoom buttons. GBrowse2 can also display interesting areas by dragging the region bar without reloading the entire page. Gene information of the selected area is also displayed, and details will be given on separate pop-up page by clicking on it.
Discussion
For a fast-paced research environment, a viewer for searching and observing data in one step is very handy. We hope our system can help researchers who are interested not only in our target polymorphism study but also in a viewer for polymorphisms for general purposes.
We chose a web-based platform for the tool because of its usability and ease of maintenance. Since its workload is very small as an input query for a viewer, a web-based platform does not have drawbacks, like other calculation applications with very large data.
Acknowledgements
This study was supported by a grant of the Korea Health 21 R&D Project, Ministry of Health and Welfare, Republic of Korea (A040002). We thank the KARE consortium for providing the original genotyping data.
Notes
Availability: The East Asian CNV database (EACDB) can be accessed at www.ircgp.com/EACNVDB.html. Some configuration and server installation should be done before the system integration. Contact yejun@catholic.ac.kr for detailed information.