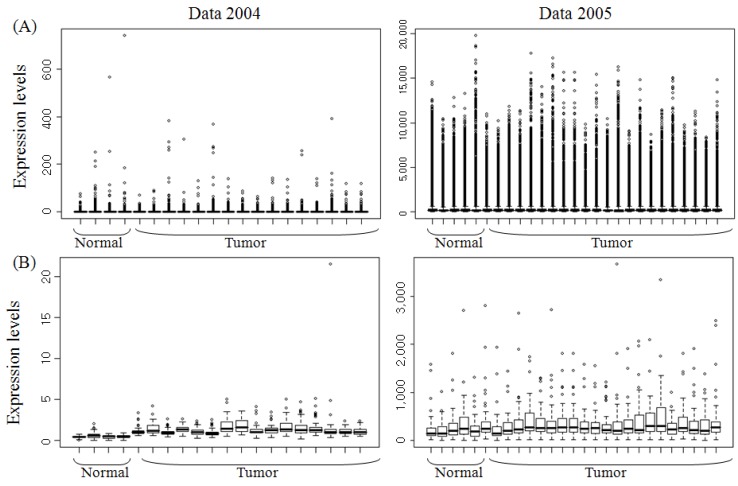
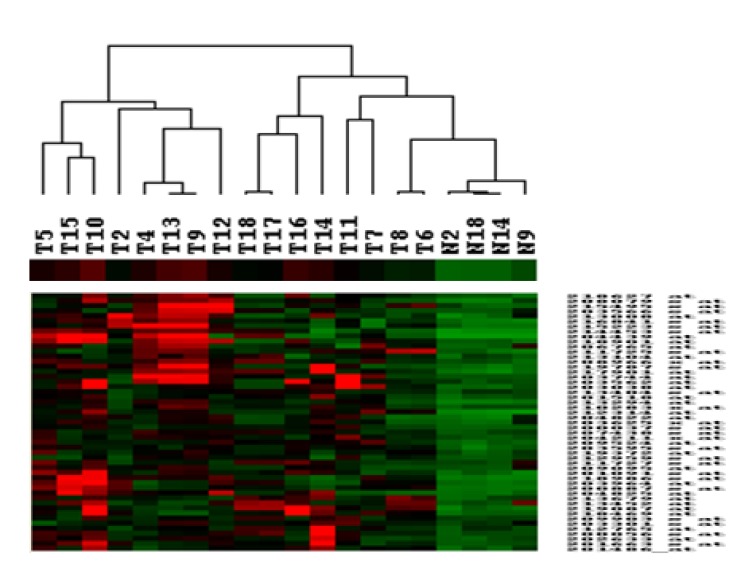
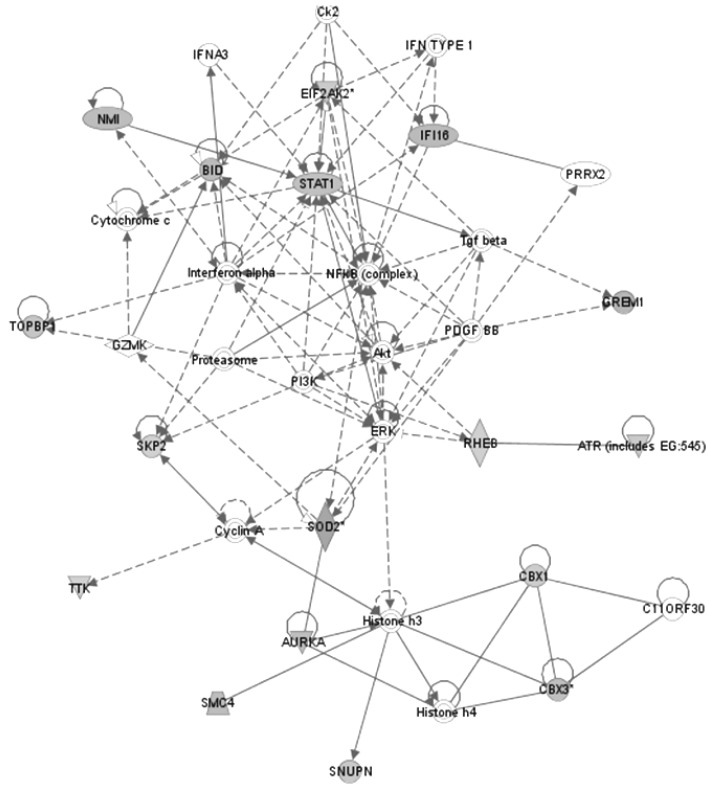
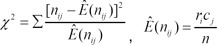1. Sparano A, Quesnelle KM, Kumar MS, Wang Y, Sylvester AJ, Feldman M,
et al. Genome-wide profiling of oral squamous cell carcinoma by array-based comparative genomic hybridization. Laryngoscope 2006;116:735ŌĆō741. PMID:
16652080.


2. Smeets SJ, Brakenhoff RH, Ylstra B, van Wieringen WN, van de Wiel MA, Leemans CR,
et al. Genetic classification of oral and oropharyngeal carcinomas identifies subgroups with a different prognosis. Cell Oncol 2009;31:291ŌĆō300. PMID:
19633365.




3. Kim KY, Ki DH, Jeung HC, Chung HC, Rha SY. Improving the prediction accuracy in classification using the combined data sets by ranks of gene expressions. BMC Bioinformatics 2008;9:283. PMID:
18554423.



4. Toruner GA, Ulger C, Alkan M, Galante AT, Rinaggio J, Wilk R,
et al. Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet Cytogenet 2004;154:27ŌĆō35. PMID:
15381369.


5. O'Donnell RK, Kupferman M, Wei SJ, Singhal S, Weber R, O'Malley B,
et al. Gene expression signature predicts lymphatic metastasis in squamous cell carcinoma of the oral cavity. Oncogene 2005;24:1244ŌĆō1251. PMID:
15558013.


6. Hiroi M, Mori K, Sakaeda Y, Shimada J, Ohmori Y. STAT1 represses hypoxia-inducible factor-1-mediated transcription. Biochem Biophys Res Commun 2009;387:806ŌĆō810. PMID:
19646959.


7. Ben-Izhak O, Akrish S, Gan S, Nagler RM. Skp2 and salivary cancer. Cancer Biol Ther 2009;8:153ŌĆō158. PMID:
19029817.


8. De Andrea M, Gioia D, Mondini M, Azzimonti B, Ren├▓ F, Pecorari G,
et al. Effects of IFI16 overexpression on the growth and doxorubicin sensitivity of head and neck squamous cell carcinoma-derived cell lines. Head Neck 2007;29:835ŌĆō844. PMID:
17510972.


10. Ye H, Wang A, Lee BS, Yu T, Sheng S, Peng T,
et al. Proteomic based identification of manganese superoxide dismutase 2 (SOD2) as a metastasis marker for oral squamous cell carcinoma. Cancer Genomics Proteomics 2008;5:85ŌĆō94. PMID:
18460737.


11. Ye H, Yu T, Temam S, Ziober BL, Wang J, Schwartz JL,
et al. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics 2008;9:69. PMID:
18254958.



12. Liu X, Yu J, Jiang L, Wang A, Shi F, Ye H,
et al. MicroRNA-222 regulates cell invasion by targeting matrix metalloproteinase 1 (MMP1) and manganese superoxide dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines. Cancer Genomics Proteomics 2009;6:131ŌĆō139. PMID:
19487542.


13. Yang ZJ, Yang G, Jiang YM, Ran YL, Yang ZH, Zhang W,
et al. Screening and sero-immunoscreening of ovarian epithelial cancer associative antigens. Zhonghua Fu Chan Ke Za Zhi 2007;42:834ŌĆō839. PMID:
18476518.

14. Hiroi M, Mori K, Sekine K, Sakaeda Y, Shimada J, Ohmori Y. Mechanisms of resistance to interferon-gamma-mediated cell growth arrest in human oral squamous carcinoma cells. J Biol Chem 2009;284:24869ŌĆō24880. PMID:
19596857.



15. Laimer K, Spizzo G, Obrist P, Gastl G, Brunhuber T, Sch├żfer G,
et al. STAT1 activation in squamous cell cancer of the oral cavity: a potential predictive marker of response to adjuvant chemotherapy. Cancer 2007;110:326ŌĆō333. PMID:
17559122.


16. Cavallo F, Astolfi A, Iezzi M, Cordero F, Lollini PL, Forni G,
et al. An integrated approach of immunogenomics and bioinformatics to identify new Tumor Associated Antigens (TAA) for mammary cancer immunological prevention. BMC Bioinformatics 2005;6(Suppl 4):S7. PMID:
16351756.



17. Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X,
et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A 2008;105:20380ŌĆō20385. PMID:
19091943.



18. Going JJ, Nixon C, Dornan ES, Boner W, Donaldson MM, Morgan IM. Aberrant expression of TopBP1 in breast cancer. Histopathology 2007;50:418ŌĆō424. PMID:
17448016.


19. Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL,
et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 2009;323:1747ŌĆō1750. PMID:
19251594.



20. Flavell JR, Baumforth KR, Wood VH, Davies GL, Wei W, Reynolds GM,
et al. Down-regulation of the TGF-beta target gene, PTPRK, by the Epstein-Barr virus encoded EBNA1 contributes to the growth and survival of Hodgkin lymphoma cells. Blood 2008;111:292ŌĆō301. PMID:
17720884.


21. van Dekken H, Vissers K, Tilanus HW, Kuo WL, Tanke HJ, Rosenberg C,
et al. Genomic array and expression analysis of frequent high-level amplifications in adenocarcinomas of the gastro-esophageal junction. Cancer Genet Cytogenet 2006;166:157ŌĆō162. PMID:
16631473.

22. Califano D, Pignata S, Pisano C, Greggi S, Laurelli G, Losito NS,
et al. FEZ1/LZTS1 protein expression in ovarian cancer. J Cell Physiol 2010;222:382ŌĆō386. PMID:
19885841.


23. Chen L, Zhu Z, Sun X, Dong XY, Wei J, Gu F,
et al. Down-regulation of tumor suppressor gene FEZ1/LZTS1 in breast carcinoma involves promoter methylation and associates with metastasis. Breast Cancer Res Treat 2009;116:471ŌĆō478. PMID:
18686028.



24. Fabris C, Basso D, Del Favero G, Meggiato T, Piccoli A, Angonese C,
et al. Renal handling of amylase and immunoreactive trypsin in pancreatic cancer and chronic pancreatitis. Clin Physiol Biochem 1990;8:30ŌĆō37. PMID:
1691065.

25. Shintani S, Li C, Mihara M, Hino S, Nakashiro K, Hamakawa H. Skp2 and Jab1 expression are associated with inverse expression of p27(KIP1) and poor prognosis in oral squamous cell carcinomas. Oncology 2003;65:355ŌĆō362. PMID:
14707456.


26. Hayes DC, Secrist H, Bangur CS, Wang T, Zhang X, Harlan D,
et al. Multigene real-time PCR detection of circulating tumor cells in peripheral blood of lung cancer patients. Anticancer Res 2006;26:1567ŌĆō1575. PMID:
16619573.

27. Fillmore RA, Mitra A, Xi Y, Ju J, Scammell J, Shevde LA,
et al. Nmi (N-Myc interactor) inhibits Wnt/beta-catenin signaling and retards tumor growth. Int J Cancer 2009;125:556ŌĆō564. PMID:
19358268.


28. Quaye L, Song H, Ramus SJ, Gentry-Maharaj A, H├Ėgdall E, DiCioccio RA,
et al. Tagging single-nucleotide polymorphisms in candidate oncogenes and susceptibility to ovarian cancer. Br J Cancer 2009;100:993ŌĆō1001. PMID:
19240718.



29. Harima Y, Ikeda K, Utsunomiya K, Shiga T, Komemushi A, Kojima H,
et al. Identification of genes associated with progression and metastasis of advanced cervical cancers after radiotherapy by cDNA microarray analysis. Int J Radiat Oncol Biol Phys 2009;75:1232ŌĆō1239. PMID:
19857786.


30. Kono K, Mizukami Y, Daigo Y, Takano A, Masuda K, Yoshida K,
et al. Vaccination with multiple peptides derived from novel cancer-testis antigens can induce specific T-cell responses and clinical responses in advanced esophageal cancer. Cancer Sci 2009;100:1502ŌĆō1509. PMID:
19459850.


31. de C├Īrcer G, P├®rez de Castro I, Malumbres M. Targeting cell cycle kinases for cancer therapy. Curr Med Chem 2007;14:969ŌĆō985. PMID:
17439397.


32. Suda T, Tsunoda T, Daigo Y, Nakamura Y, Tahara H. Identification of human leukocyte antigen-A24-restricted epitope peptides derived from gene products upregulated in lung and esophageal cancers as novel targets for immunotherapy. Cancer Sci 2007 9 02 [Epub].
http://dx.doi.org/10.1111/j.1349-7006.2007.00603.x.

33. Pickard MR, Green AR, Ellis IO, Caldas C, Hedge VL, Mourtada-Maarabouni M,
et al. Dysregulated expression of Fau and MELK is associated with poor prognosis in breast cancer. Breast Cancer Res 2009;11:R60. PMID:
19671159.




34. Kappadakunnel M, Eskin A, Dong J, Nelson SF, Mischel PS, Liau LM,
et al. Stem cell associated gene expression in glioblastoma multiforme: relationship to survival and the subventricular zone. J Neurooncol 2010;96:359ŌĆō367. PMID:
19655089.



35. Gałeza-Kulik M, Zebracka J, Szpak-Ulczok S, Czarniecka AK, Kukulska A, Gubala E,
et al. Expression of selected genes involved in transport of ions in papillary thyroid carcinoma. Endokrynol Pol 2006;57(Suppl A):26ŌĆō31. PMID:
17091453.

36. Torchia EC, Chen Y, Sheng H, Katayama H, Fitzpatrick J, Brinkley WR,
et al. A genetic variant of Aurora kinase A promotes genomic instability leading to highly malignant skin tumors. Cancer Res 2009;69:7207ŌĆō7215. PMID:
19738056.



37. Chen J, Etzel CJ, Amos CI, Zhang Q, Viscofsky N, Lindor NM,
et al. Genetic variants in the cell cycle control pathways contribute to early onset colorectal cancer in Lynch syndrome. Cancer Causes Control 2009;20:1769ŌĆō1777. PMID:
19690970.



38. Kaestner P, Stolz A, Bastians H. Determinants for the efficiency of anticancer drugs targeting either Aurora-A or Aurora-B kinases in human colon carcinoma cells. Mol Cancer Ther 2009;8:2046ŌĆō2056. PMID:
19584233.


39. Alimirah F, Chen J, Davis FJ, Choubey D. IFI16 in human prostate cancer. Mol Cancer Res 2007;5:251ŌĆō259. PMID:
17339605.


40. Zhang Y, Howell RD, Alfonso DT, Yu J, Kong L, Wittig JC,
et al. IFI16 inhibits tumorigenicity and cell proliferation of bone and cartilage tumor cells. Front Biosci 2007;12:4855ŌĆō4863. PMID:
17569615.


41. Ortega-Paino E, Fransson J, Ek S, Borrebaeck CA. Functionally associated targets in mantle cell lymphoma as defined by DNA microarrays and RNA interference. Blood 2008;111:1617ŌĆō1624. PMID:
18024791.


42. Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R,
et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet 2009;41:585ŌĆō590. PMID:
19330027.



43. Zhi G, Wilson JB, Chen X, Krause DS, Xiao Y, Jones NJ,
et al. Fanconi anemia complementation group FANCD2 protein serine 331 phosphorylation is important for fanconi anemia pathway function and BRCA2 interaction. Cancer Res 2009;69:8775ŌĆō8783. PMID:
19861535.



44. Barroso E, Pita G, Arias JI, Menendez P, Zamora P, Blanco M,
et al. The Fanconi anemia family of genes and its correlation with breast cancer susceptibility and breast cancer features. Breast Cancer Res Treat 2009;118:655ŌĆō660. PMID:
19536649.


45. Lee ES, Son DS, Kim SH, Lee J, Jo J, Han J,
et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res 2008;14:7397ŌĆō7404. PMID:
19010856.


46. Skrzycki M, Majewska M, Podsiad M, Czeczot H. Expression and activity of superoxide dismutase isoenzymes in colorectal cancer. Acta Biochim Pol 2009;56:663ŌĆō670. PMID:
19902052.


47. Olson SH, Carlson MD, Ostrer H, Harlap S, Stone A, Winters M,
et al. Genetic variants in SOD2, MPO, and NQO1, and risk of ovarian cancer. Gynecol Oncol 2004;93:615ŌĆō620. PMID:
15196853.


48. Lorch JH, Thomas TO, Schmoll HJ. Bortezomib inhibits cell-cell adhesion and cell migration and enhances epidermal growth factor receptor inhibitor-induced cell death in squamous cell cancer. Cancer Res 2007;67:727ŌĆō734. PMID:
17234784.


49. Lorch JH, Klessner J, Park JK, Getsios S, Wu YL, Stack MS,
et al. Epidermal growth factor receptor inhibition promotes desmosome assembly and strengthens intercellular adhesion in squamous cell carcinoma cells. J Biol Chem 2004;279:37191ŌĆō37200. PMID:
15205458.


50. Crighton D, Wilkinson S, Ryan KM. DRAM links autophagy to p53 and programmed cell death. Autophagy 2007;3:72ŌĆō74. PMID:
17102582.


51. Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR,
et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006;126:121ŌĆō134. PMID:
16839881.


52. Mackay A, Urruticoechea A, Dixon JM, Dexter T, Fenwick K, Ashworth A,
et al. Molecular response to aromatase inhibitor treatment in primary breast cancer. Breast Cancer Res 2007;9:R37. PMID:
17555561.




53. Turashvili G, Bouchal J, Baumforth K, Wei W, Dziechciarkova M, Ehrmann J,
et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer 2007;7:55. PMID:
17389037.




54. Fields AP, Justilien V. The guanine nucleotide exchange factor (GEF) Ect2 is an oncogene in human cancer. Adv Enzyme Regul 2010;50:190ŌĆō200. PMID:
19896966.



55. Boelens MC, Kok K, van der Vlies P, van der Vries G, Sietsma H, Timens W,
et al. Genomic aberrations in squamous cell lung carcinoma related to lymph node or distant metastasis. Lung Cancer 2009;66:372ŌĆō378. PMID:
19324446.


56. Hirata D, Yamabuki T, Miki D, Ito T, Tsuchiya E, Fujita M,
et al. Involvement of epithelial cell transforming sequence-2 oncoantigen in lung and esophageal cancer progression. Clin Cancer Res 2009;15:256ŌĆō266. PMID:
19118053.


58. Chen C, M├®ndez E, Houck J, Fan W, Lohavanichbutr P, Doody D,
et al. Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 2008;17:2152ŌĆō2162. PMID:
18669583.



59. Severino P, Alvares AM, Michaluart P Jr, Okamoto OK, Nunes FD, Moreira-Filho CA,
et al. Global gene expression profiling of oral cavity cancers suggests molecular heterogeneity within anatomic subsites. BMC Res Notes 2008;1:113. PMID:
19014556.



60. Gemenetzidis E, Bose A, Riaz AM, Chaplin T, Young BD, Ali M,
et al. FOXM1 upregulation is an early event in human squamous cell carcinoma and it is enhanced by nicotine during malignant transformation. PLoS One 2009;4:e4849. PMID:
19287496.



61. Knight JA, Onay UV, Wells S, Li H, Shi EJ, Andrulis IL,
et al. Genetic variants of GPX1 and SOD2 and breast cancer risk at the Ontario site of the Breast Cancer Family Registry. Cancer Epidemiol Biomarkers Prev 2004;13:146ŌĆō149. PMID:
14744747.


62. Liu K, Bellam N, Lin HY, Wang B, Stockard CR, Grizzle WE,
et al. Regulation of p53 by TopBP1: a potential mechanism for p53 inactivation in cancer. Mol Cell Biol 2009;29:2673ŌĆō2693. PMID:
19289498.



63. Vecchione A, Ishii H, Shiao YH, Trapasso F, Rugge M, Tamburrino JF,
et al. Fez1/lzts1 alterations in gastric carcinoma. Clin Cancer Res 2001;7:1546ŌĆō1552. PMID:
11410489.

64. Serrano-Fern├Īndez P, M├Čller S, Goertsches R, Fiedler H, Koczan D, Thiesen HJ,
et al. Time course transcriptomics of IFNB1b drug therapy in multiple sclerosis. Autoimmunity 2010;43:172ŌĆō178. PMID:
19883335.