|
 |
- Search
| Genomics Inform > Volume 14(1); 2016 > Article |
Abstract
Internet addiction (IA) has become a widespread and problematic phenomenon as smart devices pervade society. Moreover, internet gaming disorder leads to increases in social expenditures for both individuals and nations alike. Although the prevention and treatment of IA are getting more important, the diagnosis of IA remains problematic. Understanding the neurobiological mechanism of behavioral addictions is essential for the development of specific and effective treatments. Although there are many databases related to other addictions, a database for IA has not been developed yet. In addition, bioinformatics databases, especially genetic databases, require a high level of security and should be designed based on medical information standards. In this respect, our study proposes the OAuth standard protocol for database access authorization. The proposed IA Bioinformatics (IABio) database system is based on internet user authentication, which is a guideline for medical information standards, and uses OAuth 2.0 for access control technology. This study designed and developed the system requirements and configuration. The OAuth 2.0 protocol is expected to establish the security of personal medical information and be applied to genomic research on IA.
Information technology has developed so rapidly all over the world that smart devices have become popularized and digital contents have been diversified. As a consequence, internet addiction (IA) has become an important issue. Although the prevention and treatment of IA are more important than ever due to pervasive digital media and although measures to establish a systematic research environment and overcome addiction problems are more necessary than ever, unfortunately, these steps not progressing fast enough.
It has been known that IA leads to diverse psychosomatic problems, such as nonaggressive or oppositional behavior and hostility. In Korea, it was reported that 7% of internet users aged 5 to 54 years are at risk of IA, and the percentage keeps growing [1]. IA disorder, which has many similarities to psychopathological disorders, is on the rise and is considered a potential problem. Therefore, a diagnostic standard is required considering DNA, protein, and the characteristics of the structure of the brain.
Many studies have been conducted on genomes that affect addictions to substances, such as alcohol, and a considerable number of databases have been built up. However, a biological database of genes, proteins, and brain structures, which can be used directly for IA, is not sufficient.
To build a genome database related to IA, personal genome data, which are sensitive to handle, have to be examined, so security should be guaranteed. In addition, a medical information standard-based environment is very important in terms of security. It requires a medical information standard and access control technology for the service environment of genomic database [2,3].
Integration in the Healthcare Enterprise (IHE) [4] is providing various types of standard guidelines to control access to medical information. IHE access control guidelines include cross-domain user authentication (XUA), enterprise user authentication (EUA) and internet user authentication (IUA) [5].
IUA provides the guidelines for access to OAuth 2.0-based medical information service. OAuth 2.0 [6] is an access control method that manages several services in a group with one integrated ID. It is used by Google and Facebook. Therefore, it provides easy scalability into medical systems and easy control of authority for access [7].
In this paper, we propose the medical standard-based IA Bioinformatics (IABio) database platform service, which can integrate the data on genes, proteins, and brain structure image data in internet and game addiction into one database. IABio follows medical standard-based guidelines and controls access to database using OAuth 2.0. Therefore, using IABio can provide a medical standard-based service environment and will allow various integrated research studies with an access control method that enables interchange with other clinical data.
The present study is composed as follows. Relevant studies and literature on this topic are described in section II. In section III, IABio is designed. In section IV, the designed IABio is explained. Our study closes with conclusions in section V.
A genome database has many merits, such as predicting the occurrence of disease and thus preventing and swiftly diagnosing it, which leads to early treatment; determining the prognosis accurately by selecting the treatment of a certain gene; and evaluating the effectiveness of treatment.
It has been difficult to verify the connection between addiction and genes in the past, but some attempts have been made to identify the DNA responsible for addiction in the last decade. The findings say that when a genetic mutation occurs in a certain region of dopamine receptor in a human brain, dopamine receptor fails to function properly and thus can have superficial behavioral inclinations, such as addiction to smoking, alcohol, or drugs [8].
According to those studies, when genetic mutation occurs in a certain region of dopamine receptor in a human brain, dopamine receptor fails to function properly, and when the mutated genes make contact with a substance, they secrete acetylcholine, which stimulates pleasure, which leads to addiction. The genes known to be related to addiction are DRD2 Tag 1A, DRD2 Tag 1B, and so on. A German psychobiologic research team examined and compared 132 internet addicts and 711 normal people to find the cause of addiction and found that most of the addicts had a mutated gene called CHRNA4NA4 [9].
It is necessary that a database be built for addiction and that a service provision environment be established for studies regarding IA. Therefore, an integrated database is required to support genome research and analysis.
Genes, proteins, and neuroimages require tight security, because they are very sensitive information in terms of personal information privacy. To use such information, medical standards should be complied and proper guidelines should be used. Our study uses the IUA method by IHE, which provides medical standard guidelines [4,5].
IUA is an IHE profile based on access control with token-based authentication and provides access authority to medical information services and guidelines for user authentication. The IUA guideline is seen in Fig. 1.
It can grant only necessary authorities to other users according to database policy and can also provide other services in a single server, which means that it has an excellent scalability. In addition, when authorization is obtained from one authorization server, it allows the authority to use the information related to a certain user. For an authorization client to use the resource, it is necessary to obtain an authorization token from the authorization server. Authorization token holders need to check the terms of validity before using it. In case the term expires, they need to get new authorization tokens. A database environment is suitable for using other data, such as genes, proteins, and brain images, and can protect patients' data systematically. In this study, IUA is used to build a database applied with a medical information standard and OAuth 2.0 to be applied to standard IUA guidelines.
OAuth 2.0 is a protocol with authorization function to control and manage access to web services. The HTTP RESTful method is used to perform access control using an authorization token and to ask an authorization server for resources [10]. HTTP RESTful is a method to request services through a URL. Because it uses data messages in the form of JavaScript Object Notation (JSON), which is small in size, it is mainly used for mobile services at present [11]. OAuth 2.0 uses Transport Layer Security (TLS) to give and take authorization tokens and messages. There are four authorization methods. Because the authorization method differs by scenario, load in a server should be considered. It should support scalability for a large service. In addition, the service provider and authorization server are completely separate; so, server multiplexing and authorization server multiplexing are possible.
There are four authorization methods: authorization code grant (ACG), implicit grant (IG), resource owner password credentials grant (ROPCG), and client credential grant (CCG). ACG and IG are re-direction methods and should interact with resource owners and receive requests. ROPCG is an authorization method in which a client receives a resource owner's authorization information directly and relays it to the authorization server. CCG can provide requests only with an authorization token.
Complicated methods, such as ACG and IG, are not needed in a database environment for analysis, because the level of trust between users and providers is high. In addition, CCG can not be used, because resources have to be used. Therefore, ROPCG is the most appropriate method for IABio.
The IABio platform service can be applied with medical information security guidelines; can provide a portal service; and can collect and analyze databases related to IA. To design the proposed service model, our study explains the analysis of requirements, the structure of the platform service, the contents of the service, and the user authorization method.
It is necessary to analyze the complicated requirements of the system to design and develop the IABio platform service. Because security is very important to a medical information system, it requires user identification. In addition, the data exchange method, integrated data composition, and services should be specified.
It is needed to analyze patients' genomic data concerning IA for treatment and prevention and to provide them with customized service. To do so, an integrated data analysis system is required and should work with all genomic information. Users with access to IABio are classified into gene researcher, protein researcher, brain imaging researcher, senior researcher for integrated work, and server manager. Each manager has access to a patient's genome information according to the Medical Care Act, but the researcher is limited to only 'concerned' information and can not have access to other information. However, all of the researchers can have access to the risk outcome that is yielded from the integrated analysis of all genomic information. The integrated analysis risk results do not include individual patient information but are considered new information and thus are not against the Medical Care Act. Therefore, each researcher can search, enter data, and modify genomic data he manages and can search out integrated analysis outcomes.
To subdivide access from each researcher, access control is needed or else the server manager can not even read the genomic data. Therefore, a server manager can only control information that the genome manager has access to. To do so, the authorization server and IABio server have to be separated physically. Medical information standard-based guidelines and access control are necessary for authorization and approval.
Biological data (gene, protein, and brain structure image) have different identifier values. However, if several pieces of genomic data for one patient have more than one identifier, it is against data integrity, because information for one patient is duplicated. In addition, because genome information entered by each data manager is created in different systems, an identification system is needed.
Access to an integrated database requires an accessible service and web portal service in which the authorization and approval procedure has been established, which is necessary to search data related to a genome. Therefore, it is necessary to provide an integrated service using a security system suitable for medical standards. Accordingly, a web portal is designed using OAuth 2.0.
IA risk factors should be provided through integrated analysis based on data entered by each researcher and user. Integrated information should be provided for single nucleotide polymorphisms (SNP) and protein markers related to IA. Also, the accuracy of risk factors should advance enough to utilize a diagnostic index. After risk factors that are highly relevant to IA are determined through the integrated analysis of several areas in the genome, a risk evaluation score for the respective factor should be provided.
To compose such a sophisticated system, our study uses medical standard guidelines in designing the genome database platform.
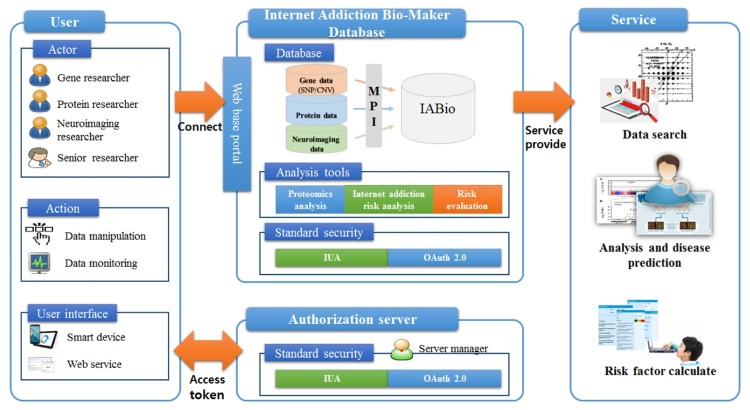
Aiming to provide a genome database platform for IA, our study designs the IABio platform service structure in consideration of medical standard security and requirements as shown in Fig. 2.
By relevance to the genome, the users are divided into gene researcher, protein researcher, brain imaging researcher, and senior researcher. Each user can operate and monitor accessible data by his/her authority. In addition, the users can connect to it through a smart device or web service.
For users to have access to IABio, they should be issued authorization tokens from the authorization server. The authorization token functions to identify researchers using genome information and is physically separated from the IABio platform. When using such an authorization token, it is possible to access IABio. When an authorization token is not valid or when a user is not registered, the token should be issued or re-issued by the authorization server. Our study complies with IUA medical standard guidelines for access, uses OAuth 2.0, and composes the security system.
IABio is accessible through a web portal. Genome data entered by each researcher are entered in a database in an agreeable format to master patient index (MPI) structure. Because gene, protein, and brain imaging data are all in different data formats, a method is necessary to integrate them. Therefore, MPI is used. It allows scalability in connection with different clinical data. MPI configures the data format in a way to identify them, develops them into a hash-type algorithm, creates an identification system, and maintains data integrity. In an MPI-based identification system, each manager does not know the password structure. Therefore, researchers can not have access to different data. The security system of the proposed IABio platform has pre-defined password keys with the authorization server and performs the authorization and approval procedure by the user. The contents of MPI are not described here, because they are not part of the aim of this study.
Three analyses are performed to provide services from IABio. The first is proteomics analysis. Proteomics is a study concerned with the structure and analysis of proteins occurring in living organisms. It aims to analyze proteins on a great scale; write a map of functional interrelations; and demonstrate a certain protein and the gene that creates it through structural analysis. The second analysis is IA risk factor analysis. SNPs and protein markers related to IA yield risk factors for IA. In addition, it analyzes clinical data, such as survey, patient interview, functional magnetic resonance imaging, and brain imaging, in addition to a genome/protein index, in integrated manner, finally creating a diagnostic index. The third analysis is risk evaluation analysis. When risk factors highly relevant to IA are determined by integrated analysis of genome data, it generates an algorithm to provide a risk evaluation score for the respective factor. In this study, the contents of genome analyses are not described, because they are separate areas of study.
Because IABio service model proposed in our study can ensure safe access to information entered by users and can provide integrated data analysis service. It can be applied to several platforms as well as studies related to IA.
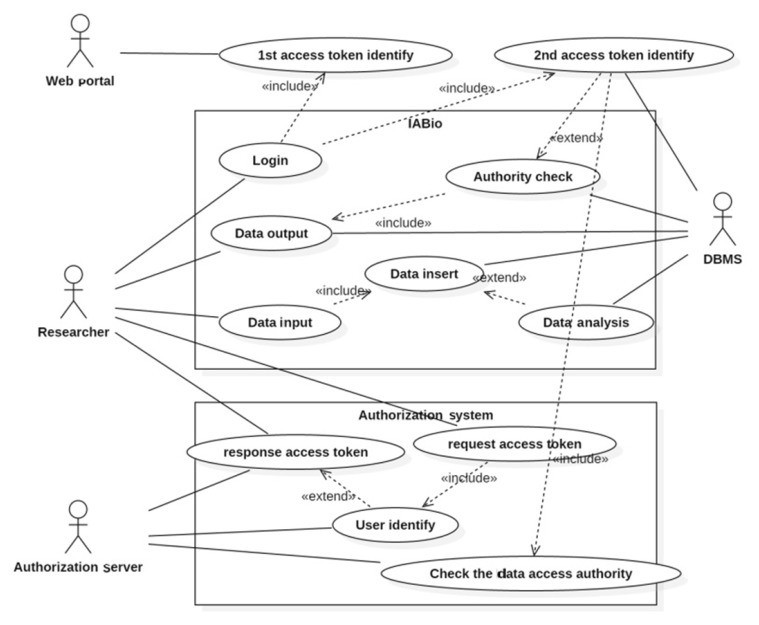
Fig. 3 shows the authorization processing procedure of the token method and the service contents that each user is provided in the use case diagram.
Four actors consist of a researcher, database, web portal, and authorization server. A researcher accesses the authorization system to have access to the IABio platform and requests an access token. The authorization server confirms the legitimacy of the access by the user and issues the right access token for his/her authority. Then, the researcher acquires the access token.
The researcher proceeds to access the IABio platform through a portal. The web portal and database identify the access token. When the login is done properly, the user can input and output data. Data can be entered according to information related to a concerned genome. Genome input data are analyzed, and the analyzed information is saved.
The researcher requests data output to monitor the analysis results and related data. Token authorization contents are analyzed for a second time to check if the researcher is legitimate for data confirmation before the analysis results and entered information are retrieved.
In this study, the IUA guideline [5] and OAuth 2.0 [6] are used to design and realize IABio. Researchers should obtain access authorization to use IABio and the concerned data. The server manager who controls the authorization server should not have access to the database in which patient information is saved. Therefore, the IABio server should be physically separated from the authorization server and provide only authorization and authority. This kind of environment is suitable for the IUA guideline. In this study, the authorization method uses the OAuth 2.0-based ROPCG method and is designed in a communication diagram, as in Fig. 4.
Our study proposes an authorization method based on the 2-step access authorization of IABio, along with resource owner server (web portal) authorization. Each researcher (client) should be assigned an authorization token from the authorization server to get access to the portal. After obtaining the authorization token, the researcher has access to the portal and proceeds with the first authorization. Next, the researcher gets the second authorization in IABio to use the service. Because security authorization is strictly designed in double walls, it can avoid weaknesses to phishing scams and impersonation attacks.
The first step of the authorization procedure is to check if it is possible to identify users' personal information. The second authorization step proceeds with authorization for database access one more time.
Our study uses this 2-step authorization in designing and developing IABio.
Fig. 5 shows the system environment in which the IABio system is composed with OAuth 2.0 of a medical information standard environment.
The system environment has the structure of a hardware and development environment. Two servers are used: IABio server and authorization server. HP ProLiant Micro Server Gen8 is used to design the IABio database, and it consists of 1TB SSD, 2TB HDD, and 12GB RAM. To compose the development environment, Apache 2.2 is used as server software to configure the Tomcat Server. For the web service, PHP 5.2 is used to supplement the functions of existing HTTP language. MySQL 5.1 is used to compose the Tomcat server and PHP, which can interwork with the database. APM (Apache, PHP, MySQL) is mutually complementary and flexible for interworking. Furthermore, it is compatible with all operating systems, such as Unix, Linux, and Windows.
The authorization server uses Naver application programming interface (API), which supports OAuth 2.0 [12]. Since Naver API provides a security server in itself, it is cost-effective. In addition, it is possible to customize so as to receive an authorization token by using Naver ID. Currently, most domestic web service providers related to authorization use OAuth 2.0 of Naver API. Therefore, it can ensure security. JSON method is employed for the exchange of messages between a portal and database server and authorization server. The JSON method enables message exchange with shorter codes than the existing Extensible Markup Language (XML) method. Therefore, it is faster in speed and can reduce load.
For security protocol, IUA guidelines and OAuth 2.0 are used. PHP 2.2 is used to realize OAuth 2.0. The JSON method can transmit authorization tokens and protect users' personal information.
The database module is structured to have access to genome data. The authorization token method should be used to have access to a database module. The Advanced Encryption Standard (AES) 256 algorithm is used to enhance the level of security. AES 256 is a symmetric key algorithm that supports block passwords. AES 256 is the most reliable symmetric key algorithm among any developed thus far. Its method uses key values used between a user and service. A user can access the web portal and database without concerned keys. PHP 2.2 is used for the module to have access to and operate IABio. In addition, MPI is used to ensure the integrity of the genome database. It can support interworking between data. Therefore, data integration is possible. Furthermore, because MPI uses Hash 256, each user can confirm which data are interworked. The Bio-Marker database is composed so as to receive the entry of gene data, protein data, and brain imaging data.
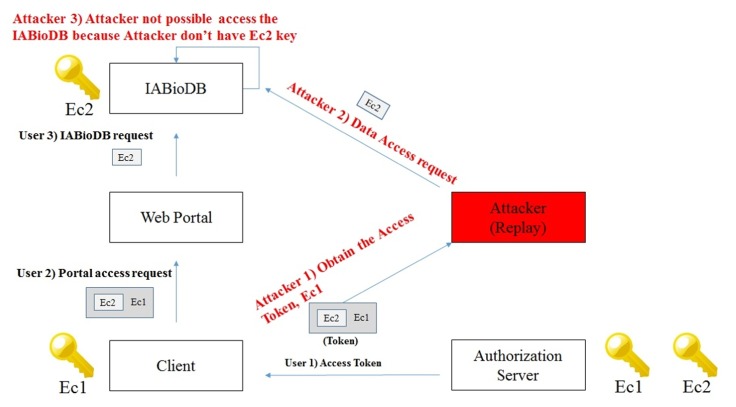
OAuth 2.0 has a weakness for vulnerable replay attacks. In this study, a replay attack can solve problems by using the 2-step access authorization. Replay attack and defense scenarios are shown in Fig. 6.
An attacker is disguised as a client to obtain an EC1 key (with access token) when passed during the communication between the authorization server and client. The attacker, disguised as a client, attempts to perform a replay attack obtained using the access token. In this case, the attacker has access to the web portal using the EC1 key (with access token) shared with the authentication server. However, the EC2 key is needed to access IABio database. An attacker who does not have the EC2 key can not access the data, because the attacker can not analyze contents in the access token of EC2. Thus, an attacker can not access the IABio database, because the attacker does not have the EC2 key. EC1 and EC2 can be obtained only from the pre-consultation process.
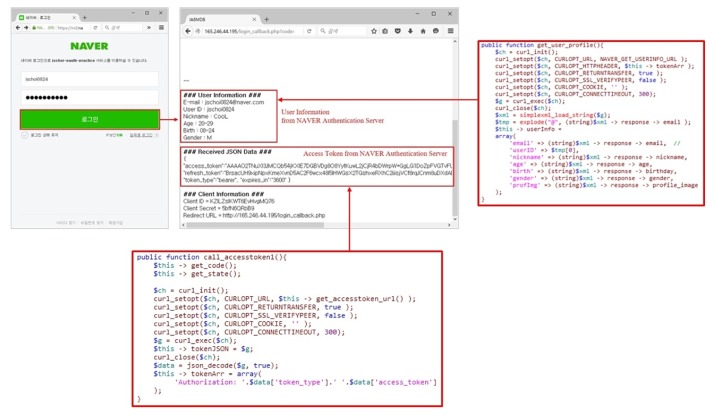
The IABio platform uses the authorization server provided in Naver API [12]. Therefore, it can acquire an authorization token in Naver and can have access to the portal by using it. To access the authorization server, only a pre-authorized Naver ID can be used. Therefore, a token can be issued or re-issued only when having client information. The left side of Fig. 7 shows the page to access the web portal, and the right side is the page to receive authorization tokens through Naver.
As seen in Fig. 8, login is done to the authorization server with client information. Then, it is possible to have access to the web portal of IABio. The right side of Fig. 8 shows the contents of personal information and the authorization token received from Naver. The authorization token is transmitted in the encryption of AES 256. The contents are those of decoded authorization tokens.
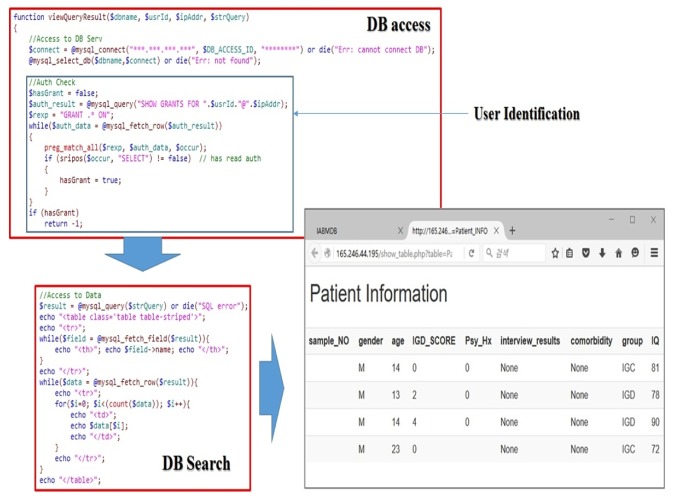
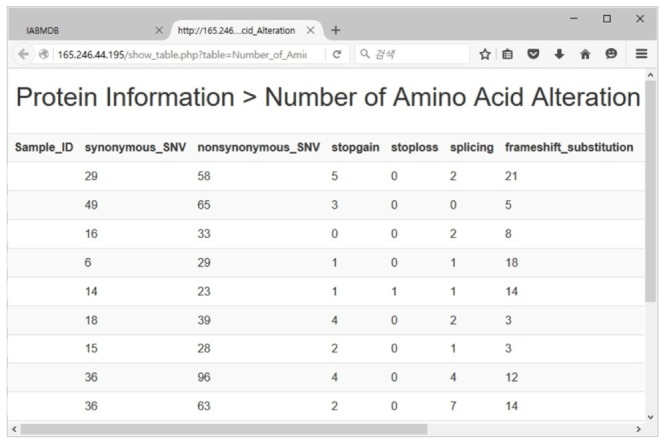
Figs. 9 and 10 are output results made by query from the data saved in the genome database. It is a screenshot of sample data, not actual clinical data. Actual data can not be printed out due to the Medical Care Act. Fig. 9 shows the screen that confirms the patient's basic information. Only personnel who are authorized for access to it can read it. Fig. 10 is a screenshot that confirms protein data. Only the personnel who are authorized for access to protein data can read it.
IA is a kind of illness in which the occurrence increases along with the development of IT technology. Although it is urgent to have an analysis database of IA, it is not sufficient now. Because the database needed to analyze medical information handles very sensitive personal data, security is very important. Our study proposed the IABio platform service using OAuth in a medical information standardbased environment. The proposed IABio uses IUA as a medical standard guideline and complies with medical information standards. Also, it uses OAuth 2.0, which is an access control technology. To realize IABio, it is necessary to analyze the complicated requirements of a medical information system and to design the system from the perspective of security. In addition, our study developed a prototype IABio system and tested if it could be applied with a security system. As a result, it was found that each user had a different environment in which the researcher needed to access a database. Therefore, it is expected that the proposed IABio can be actively used for studies of IA.
As follow-up research, solutions to phishing scams and re-use attack, to which OAuth 2.0 is vulnerable, will be examined. Also, an index searching method will be studied to quickly search genome information related to IA.
Acknowledgments
This work was supported by a National Research Foundation of Korea (NRF) grant, funded by the Korean government (MSIP:Ministry of Science, ICT and Future Planning) (NRF-2015M3C7A1064796).
References
1. National Information Society Agency. Ministry of Science. ICT and future planning: 2013 internet addiction research on the actual condition. Daegu: National Information Society Agency, 2014.
2. Beimel D, Peleg M. The context and the SitBAC models for privacy preservation: an experimental comparison of model comprehension and synthesis. IEEE Trans Knowl Data Eng 2010;22:1475–1488.

3. Jeong CW, Kim WH, Yoon KH, Joo SC. Medical information dynamic access system in smart mobile environments. J Korean Soc Internet Inf 2015;16:47–55.


4. IHE Technical Frameworks. Oak Brook: IHE International, Inc., 2015. Accessed 2015 Mar 17. Available from: http://www.ihe.net/technical_frameworks.
5. ITI Technical Committee. IHE IT infrastructure technical framework supplement: internet user authorization (IUA). Oak Brook: IHE International, Inc., 2015. Accessed 2015 Mar 17. Available from: http://www.ihe.net/uploadedFiles/Documents/ITI/IHE_ITI_Suppl_IUA.pdf.
6. Hardt D. The OAuth 2.0 authorization framework. IETF RFC 6749. IETF Tools, 2012.
7. Kang SU. An access control management based on medical standard in healthcare information services. Suwon: Ajou University, 2015. M.S. Thesis.
8. Rosell DR, Zaluda LC, McClure MM, Perez-Rodriguez MM, Strike KS, Barch DM, et al. Effects of the D1 dopamine receptor agonist dihydrexidine (DAR-0100A) on working memory in schizotypal personality disorder. Neuropsychopharmacology 2015;40:446–453. PMID: 25074637.


9. Montag C, Kirsch P, Sauer C, Markett S, Reuter M. The role of the CHRNA4 gene in internet addiction: a case-control study. J Addict Med 2012;6:191–195. PMID: 22722381.


10. Rodriguez A. RESTful web services: the basics. Armonk: IBM, 2008. Accessed 2015 Mar 17. Available from: http://www.gregbulla.com/TechStuff/Docs/ws-restful-pdf.pdf.
11. Wagh K, Thool R. A Comparative study of SOAP vs REST web services provisioning techniques for mobile host. J Inf Eng Appl 2012;2:12–16.
12. Naver open API. Seongnam: Naver Corp., 2015. Accessed 2015 Mar 17. Available from: http://developer.naver.com/wiki/pages/OpenAPI.
Fig. 4
Communication diagram of authentication method with resource owner password credentials grant [6]. IABio, Internet Addiction Bioinformatics.
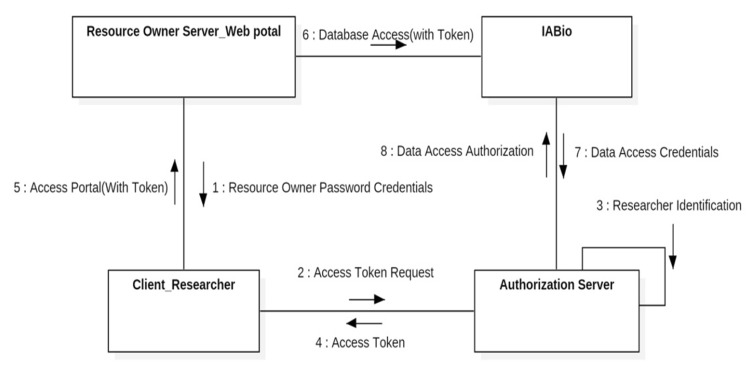
Fig. 5
Internet Addiction Bioinformatics (IABio) system environment. MPI, master patient index; JSON, JavaScript Object Notation; API, application programming interface.
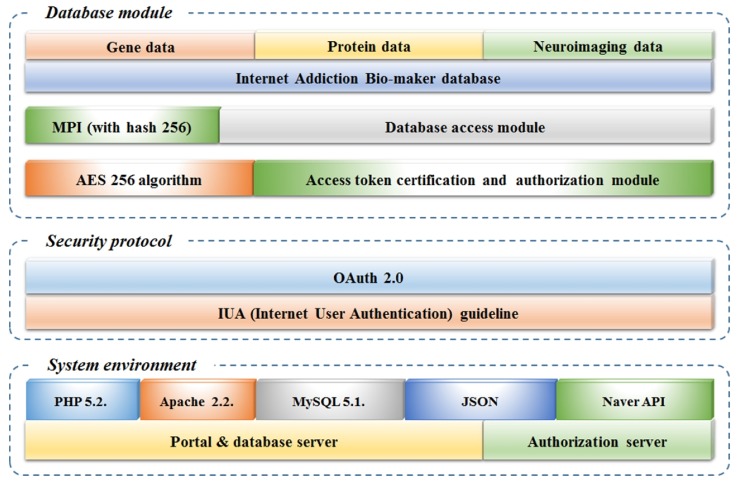
Fig. 7
Authorization server page (left and right images indicate web portal login page and Naver login page for access token, respectively) [12]. IABio DB, Internet Addiction Bioinformatics database.
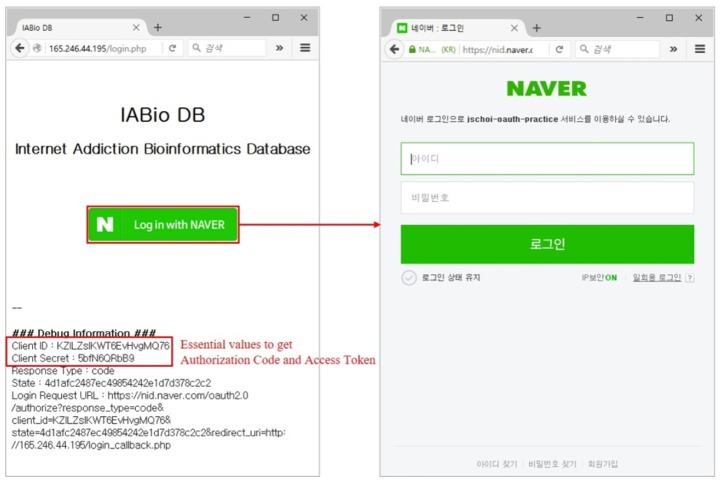
Fig. 8
IABioDB web portal (left and right images indicate Naver login page and IABioDB web portal access, respectively) [12].
- TOOLS
-
METRICS

-
- 1 Crossref
- 0 Scopus
- 7,473 View
- 82 Download
- Related articles in GNI