1. Ueki M, Cordell HJ. Improved statistics for genome-wide interaction analysis. PLoS Genet 2012;8:e1002625. PMID:
22496670.



2. Won S, Kwon MS, Mattheisen M, Park S, Park C, Kihara D,
et al. Efficient strategy for detecting gene x gene joint action and its application in schizophrenia. Genet Epidemiol 2014;38:60ŌĆō71. PMID:
24272960.


3. Wang X, Elston RC, Zhu X. Statistical interaction in human genetics: how should we model it if we are looking for biological interaction? Nat Rev Genet 2011;12:74. PMID:
21102529.



4. Hu JK, Wang X, Wang P. Testing gene-gene interactions in genome wide association studies. Genet Epidemiol 2014;38:123ŌĆō134. PMID:
24431225.



5. Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF,
et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet 2001;69:138ŌĆō147. PMID:
11404819.



6. Zhang Y, Liu JS. Bayesian inference of epistatic interactions in case-control studies. Nat Genet 2007;39:1167ŌĆō1173. PMID:
17721534.


7. Schwarz DF, K├Čnig IR, Ziegler A. On safari to Random Jungle: a fast implementation of Random Forests for high-dimensional data. Bioinformatics 2010;26:1752ŌĆō1758. PMID:
20505004.



8. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D,
et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559ŌĆō575. PMID:
17701901.



9. Wan X, Yang C, Yang Q, Xue H, Fan X, Tang NL,
et al. BOOST: a fast approach to detecting gene-gene interactions in genome-wide case-control studies. Am J Hum Genet 2010;87:325ŌĆō340. PMID:
20817139.



10. Yung LS, Yang C, Wan X, Yu W. GBOOST: a GPU-based tool for detecting gene-gene interactions in genome-wide case control studies. Bioinformatics 2011;27:1309ŌĆō1310. PMID:
21372087.



11. Ko SH, Kim SR, Kim DJ, Oh SJ, Lee HJ, Shim KH,
et al. 2011 clinical practice guidelines for type 2 diabetes in Korea. Diabetes Metab J 2011;35:431ŌĆō436. PMID:
22111032.



12. Park SE, Lee WY, Oh KW, Baek KH, Yoon KH, Kang MI,
et al. Impact of common type 2 diabetes risk gene variants on future type 2 diabetes in the non-diabetic population in Korea. J Hum Genet 2012;57:265ŌĆō268. PMID:
22377714.


13. Park KS. The search for genetic risk factors of type 2 diabetes mellitus. Diabetes Metab J 2011;35:12ŌĆō22. PMID:
21537408.



14. Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ,
et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet 2009;41:527ŌĆō534. PMID:
19396169.


15. Agresti A. Categorical Data Analysis. 2nd ed. New York: Wiley-Interscience, 2002.

16. Matsuda H. Physical nature of higher-order mutual information: intrinsic correlations and frustration. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics 2000;62(3 Pt A):3096ŌĆō3102. PMID:
11088803.


17. Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457ŌĆō470. PMID:
22384356.



18. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904ŌĆō909. PMID:
16862161.


19. Ross KA. Evidence for somatic gene conversion and deletion in bipolar disorder, Crohn's disease, coronary artery disease, hypertension, rheumatoid arthritis, type-1 diabetes, and type-2 diabetes. BMC Med 2011;9:12. PMID:
21291537.



 .
. be the MLE of ┬Ąijk for MH, we have
be the MLE of ┬Ąijk for MH, we have
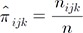 and
and  , it can be further simplified as and it can also be denoted by Kullback-Leibler's [9] form as
, it can be further simplified as and it can also be denoted by Kullback-Leibler's [9] form as
 is known to be an upper bound of
is known to be an upper bound of  , and they are almost identical if they are larger than 25. The default value of the threshold Žä in BOOST is 30, and its corresponding p-value is 4.89 ├Ś 10ŌłÆ6 [9].
, and they are almost identical if they are larger than 25. The default value of the threshold Žä in BOOST is 30, and its corresponding p-value is 4.89 ├Ś 10ŌłÆ6 [9]. >Žä, the interaction will be considered in the next testing stage; otherwise, it will be discarded from the analyses. All of the non-significant SNP pairs would be filtered out in this manner in the screening stage.
>Žä, the interaction will be considered in the next testing stage; otherwise, it will be discarded from the analyses. All of the non-significant SNP pairs would be filtered out in this manner in the screening stage.







