 |
 |
- Search
| Genomics Inform > Volume 13(3); 2015 > Article |
Abstract
MicroRNAs (miRNAs) have been demonstrated to play an important role in carcinogenesis. Previous studies revealed that miRNAs are present in human plasma in a remarkably stable form that is protected from endogenous RNase activity. In this study, we measured the plasma expression levels of three miRNAs (miR-21, miR-27a, and miR-155) to investigate the usefulness of miRNAs for gastric cancer detection. We initially examined plasma miRNA expression levels in a screening cohort consisting of 15 patients with gastric cancer and 15 healthy controls from Korean population, using TaqMan quantitative real-time polymerase chain reaction. We observed that the expression level of miR-27a was significantly higher in patients with gastric cancer than in healthy controls, whereas the miR-21 and miR-155a expression levels were not significantly higher in the patients with gastric cancer. Therefore, we further validated the miR-27a expression level in 73 paired gastric cancer tissues and in a validation plasma cohort from 35 patients with gastric cancer and 35 healthy controls. In both the gastric cancer tissues and the validation plasma cohort, the miR-27a expression levels were significantly higher in patients with gastric cancer. Receiver-operator characteristic (ROC) analysis of the validation cohort, revealed an area under the ROC curve value of 0.70 with 75% sensitivity and 56% specificity in discriminating gastric cancer. Thus, the miR-27a expression level in plasma could be a useful biomarker for the diagnosis and/or prognosis of gastric cancer.
In the past decade, gastric cancer has become of the most common malignancies worldwide. Despite significant improvements in screening and treatment technologies for gastric cancer, it remains the second most common cause of cancer-related deaths [1]. An accurate biomarker for detection of gastric cancer may reduce the cancer-related mortality. Although conventional strategies for blood-based biomarker discovery have shown promise, the development of clinically validated cancer detection markers remains an unmet challenge for many common human cancers [2]. New approaches that can complement and improve on current strategies for cancer detection are urgently needed.
MicroRNAs (miRNAs) are a subset of non-coding RNA molecules (approximately 22 nucleotide in length) [3] that negatively regulate the protein expression of specific mRNAs by imperfectly base pairing together, resulting in protein translational repression of the target gene [4]. Growing evidence has shown that miRNAs are involved in a variety of biological processes, including cell proliferation, differentiation, and apoptosis [5,6,7]. Studies of miRNAs have been extended to many kinds of tumors [8] because altered expression of miRNAs has been demonstrated to play an important role in carcinogenesis, either by oncogenic [9,10] or tumor suppressor functions [11,12].
For example, three miRNAs (miR-21, miR-27a, and miR-155) have been reported as oncogenic miRNAs. MiR-21, the most commonly up-regulated miRNA in both solid and hematological tumor tissues [8,13], directly targets tumor suppressors such as PTEN phosphatase, actin-binding protein tropomyosin I and reversion-inducing-cysteine-rich protein with Kazal motif [9,14,15]. MiR-27a is an oncogenic miRNA in gastric adenocarcinoma, targeting prohibition [16], and genetic variant of miR-27a is associated with its expression and increased risk of various cancer [17,18,19]. MiR-155 is frequently overexpressed in many cancer tissues and targets tumor protein p53 inducible nuclear protein 1 and FOXO3a [20,21].
Previously, studies revealed that tumor-associated miRNAs are present in human plasma and serum in a remarkably stable form that is protected from endogenous RNase activity [22,23], and can be readily detected in these blood fluids [24,25,26]. In addition, it was demonstrated that the oncogenic miRNA expression levels in plasma and saliva were decreased in cancer patients post-surgery [27,28,29,30]. These findings suggest that miRNAs have the potential to be useful diagnostic or prognostic biomarkers for cancer detection.
In this study, we evaluated the expression levels of these three miRNA variants in plasma samples from a screening cohort of 15 patients with gastric cancer and 15 healthy controls in Korean population. The significant miR-27a expression levels found were further validated in an additional 73 paired gastric cancer tissues and 70 plasma samples from Korean population, using quantitative real-time polymerase chain reaction (qRT-PCR).
The plasma and tissue samples (including matched non-tumor tissues) from patients with gastric cancer were retrieved from the Chungnam National University Hospital, Republic of Korea. All patients gave their written consent prior to sample collection. The acquisition of clinical material was approved by the local institution review board. These plasma samples were drawn from 35 patients (18 men, 17 women) with gastric neoplasm and 35 control individuals (18 men, 17 women) without neoplastic disease. There was no significant difference in age between the neoplastic group (51.80 ± 11.88 years) and the control group (48.94 ± 11.86 years) (Supplementary Table 1). The demographic characteristics of the subjects who provided the 73 paired tissue samples are summarized in Supplementary Table 2.
Venous blood from all subjects was collected into sterile ethylenediamine tetracetic aci-coated vials. Within 1 h, samples were centrifuged (2,000 ×g, 10 min), following which the plasma was removed, aliquoted, and stored at -80℃ until isolation of the circulating RNA. Total RNA was isolated from 300 µL of plasma using the Ambion PARIS Kit (Ambion, Austin, TX, USA) according to the manufacturer's instruction. Total RNA from tissue samples was extracted with the miRNeasy Midi Kit (Qiagen, Carlsbad, CA, USA), and finally eluted into 80 µL of elution solution according to the manufacturer's instructions. The total RNAs isolated from plasma and tissues were stored at -80℃ until further use.
The miR-21, miR-27a, and miR-155 expression levels were measured by using the TaqMan qRT-PCR analysis system (Applied Biosystem, Foster City, CA, USA). The expression level of each miRNA was normalized to U6snRNA. In brief, this assay entailed a 2-step qRT-PCR of miRNA of 19-25 nucleotides, priming with a stem-loop primer onto a longer cDNA that is amenable to amplification, and quantification by a TaqMan-based real-time PCR. Ct was the number of cycles at which the fluorescence signal passed the appropriate threshold. Expression levels were determined by using the delta Ct (ΔCt) method, where Ct represents the number of cycles in which the fluorescence signal passed the appropriate threshold.
For the detection of miRNA, 2 µg of total RNA extracted from tissues was reverse transcribed into cDNA using M-MLV (Promega, Madison, WI, USA) with miRNA-specific stem-loop primers, as previously described [16]. The cDNA was used for the amplification of mature miR-27a and endogenous control (U6snRNA) by real-time PCR. The real-time PCR was performed by using the SYBR Green PCR Mix (Bio-Rad, Hercules, CA, USA) with a CFX96 real-time PCR system (Bio-Rad). The PCR cycles were as follows: initial denaturation at 95℃ for 0.5 min, followed by 40 cycles at 94℃ for 0.5 min, 56℃ for 1 min, 72℃ for 0.5 min. The relative miRNA expression level was calculated using the delta Ct (ΔCt) method.
The significance of miRNA expression level difference between patients with gastric cancer and cancer-free control subjects was analyzed using Student's t-test. Determination of the receiver-operator characteristic (ROC), and the respective area under the ROC curve (AUC), and Kaplan-Meier survival analysis were performed using the R software (version 2.6.1). Results with a p-value of less than 0.05 were considered significant.
To select potential miRNAs for gastric cancer detection, we divided our samples into the 30 screening cohort and 40 validation cohort. The expression levels of three well-known oncogenic miRNAs were evaluated in 30 plasma samples consisting of 15 patients with gastric cancer and 15 healthy controls. Box plots for the quantitative fluorogenic real-time PCR values are shown in Fig. 1. The miR-21 and miR-155 expression levels were not significantly different between the neoplastic group and the control (p = 0.08 and p = 0.27) (Fig. 1A and 1C). However, the plasma miR-27a expression levels were significantly higher in the patients with gastric cancer (p = 0.002) (Fig. 1B). Although miR-21 expression level in plasma was tend to be increased in patient with gastric cancer compared to healthy control (p = 0.08), we selected miR-27a as a high priority for further analysis.
We examined the miR-27a expression level in 73 paired gastric cancer tissues to confirm the oncogenic expression of this miRNA variant. The miR-27a expression level was elevated 2.8-fold in gastric cancer tissues compared with non-tumor tissues (p = 1.96e-06) (Fig. 2A), and this miRNA was highly expressed in 61 out of 73 gastric cancer patient samples (80%). The miR-27a expression levels were highly elevated in all stages compared with non-tumor tissues (Fig. 2B). We divided the patient samples into diffuse and intestinal types, based on the Lauren classification, because gastric cancer patients with diffuse-type tumors have a much poorer prognosis than those with the intestinal type. We observed that the miR-27a expression levels were not significantly different between intestinal and diffuse types (p = 0.19; data not shown). By ROC analysis, we observed that the optimal cut-off value for discriminating between patients and healthy individuals was 0.0067 for a miR-27a expression. miR-27a had 73% sensitivity and 62% specificity in discriminating gastric cancer (Fig. 2C). Importantly, we observed a significant association between miR-27a overexpression and poor survival (p = 0.03) (Fig. 2D), suggesting that miR-27a has potential to be used as a prognostic marker for patients with gastric cancer. Taken together, the miR-27a expression pattern resembled that of an oncogenic microRNA and was associated with gastric cancer patient survival.
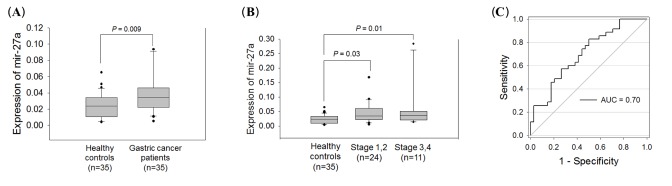
To further validate the diagnostic value of miR-27a, its expression level was measured in additional plasma samples drawn from 20 patients with gastric cancer and 20 healthy controls. The result of miR-27a expression data obtained from each 15 patients with gastric cancer and healthy control plasma samples used in screening cohort were also included in the validation cohort, because of limited sample size. The qRT-PCR results revealed that the miR-27a expression level was highly elevated in the neoplastic group (p = 0.009) (Fig. 3A). In both early (I and II) and late stages (III and IV), the miR-27a expression levels were significantly higher in the group with gastric cancer (Fig. 3B). By ROC analysis, we observed the AUC value to be 0.70 with 75% sensitivity and 56% specificity in discriminating gastric cancer based on the cut-off value of 0.031 for miR-27a expression level in plasma (Fig. 3C). Our results suggest that the plasma miR-27a expression level may have potential as a non-invasive diagnostic biomarker for gastric cancer.
A diagnostic source that would provide high specificity and sensitivity may be of enormous benefit to cancer patients, particularly if the biosource could be obtained from non-invasive materials. It was demonstrated that miRNAs are involved in multiple steps of carcinogenesis, either by oncogenic or tumor suppressor functions [31], and circulating miRNAs exist in the serum or plasma [29,32,33]. More importantly, miRNAs have been detected in human serum and plasma in stable forms because of their short hairpin structures, encapsulation by microvesicles, and association with protein complexes [22,23,34,35]. Thus, the detection of any altered expression of tumor-specific miRNAs in saliva, serum, plasma, and urine may offer a promising approach for non-invasive gastric cancer detection.
In this study, the plasma expression levels of three miRNAs (miR-21, miR-27a, and miR-155) were measured in a screening cohort, and we observed that miR-27a expression was significantly elevated in gastric cancer samples compared with healthy control samples. Our result is similar to previously reported results [36]. Next, we investigated whether plasma miR-27a could be released from gastric tumor tissues. We observed that the miR-27a expression level was similar between gastric tumor tissues and plasma, suggesting that the plasma miR-27a expression level may reflect that of the gastric tumor tissues.
miR-27a is located on chromosome 19, and has oncogenic function in gastric adenocarcinoma, by targeting prohibition [16]. This miRNA variant is associated with lymph node metastasis and is overexpressed in gastric cancer [17]. In addition, miR-27a can regulate tumor suppress genes such as SPRY2, and FBW7 [37,38]. SPRY2 can inhibit tumor growth and metastases by interfering with Ras/MAPK activation [37], and FBW7 acts as the substrate recognition component of a Skp1-Cul1-F-box-protein ubiquitin ligase that targets numerous oncoproteins for proteasomal degradation [38]. Growing evidence has suggested that miR-27a has an important role in gastric cancer development and progression.
To estimate its potential value for gastric cancer detection, we evaluated the plasma miR-27a expression level in a validation cohort consisting of 35 patients with gastric cancer and 35 healthy control samples. The sensitivity for the validation cohort was estimated as 75% and the specificity as 56%. Current diagnostic tools for gastric cancer, such as the serological markers carbohydrate antigen 19-9 and carcinoembryonic antigen have low specificity and sensitivity [36]. Our data indicates that the sensitivity and specificity of plasma miR-27a are better than that of the serological markers, although plasma miR-27a alone is not sufficient for accurate gastric cancer detection. Thus, real improvement in the accuracy of gastric cancer detection still needs to be achieved by recruiting additional plasma miRNA markers.
In conclusion, plasma miR-27a could be a useful non-invasive biomarker for gastric cancer detection. Additional studies using a large cohort of samples are needed to validate these results because our study had a small sample size.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF), grants funded by the Korea government (MSIP; No. 2011-0030049), and a KRIBB Research Initiative Grant.
Supplementary materials
Supplementary data including two tables can be found with this article online at http://www.genominfo.org/src/sm/gni-13-70-s001.pdf.
Supplementary Table 1
Clinical characteristics of the plasma samples from 35 patients with gastric cancer and 35 healthy controls
Supplementary Table 2
Clinical characteristics of the 73 paired gastric cancer tissue samples
References
1. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74–108. PMID: 15761078.


2. Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol 2006;24:971–983. PMID: 16900146.


3. Kim VN. Small RNAs: classification, biogenesis, and function. Mol Cells 2005;19:1–15. PMID: 15750334.


4. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–297. PMID: 14744438.


6. Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol 2009;27:5848–5856. PMID: 19884536.



7. Meola N, Gennarino VA, Banfi S. MicroRNAs and genetic diseases. Pathogenetics 2009;2:7. PMID: 19889204.



8. Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 2008;13:272–286. PMID: 18328430.


9. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007;133:647–658. PMID: 17681183.



10. Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res 2010;70:3119–3127. PMID: 20354188.


11. Lee KH, Chen YL, Yeh SD, Hsiao M, Lin JT, Goan YG, et al. MicroRNA-330 acts as tumor suppressor and induces apoptosis of prostate cancer cells through E2F1-mediated suppression of Akt phosphorylation. Oncogene 2009;28:3360–3370. PMID: 19597470.


12. Mishra PJ, Song B, Mishra PJ, Wang Y, Humeniuk R, Banerjee D, et al. miR-24 tumor suppressor activity is regulated independent of p53 and through a target site polymorphism. PLoS One 2009;4:e8445. PMID: 20041160.



13. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257–2261. PMID: 16461460.



14. Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem 2007;282:14328–14336. PMID: 17363372.


15. Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A 2008;105:7004–7009. PMID: 18458333.



16. Liu T, Tang H, Lang Y, Liu M, Li X. MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett 2009;273:233–242. PMID: 18789835.


17. Sun Q, Gu H, Zeng Y, Xia Y, Wang Y, Jing Y, et al. Hsa-mir-27a genetic variant contributes to gastric cancer susceptibility through affecting miR-27a and target gene expression. Cancer Sci 2010;101:2241–2247. PMID: 20666778.


18. Nikolić Z, Savić Pavićević D, Vučić N, Cidilko S, Filipović N, Cerović S, et al. Assessment of association between genetic variants in microRNA genes hsa-miR-499, hsa-miR-196a2 and hsa-miR-27a and prostate cancer risk in Serbian population. Exp Mol Pathol 2015;99:145–150. PMID: 26112096.


19. Shi D, Li P, Ma L, Zhong D, Chu H, Yan F, et al. A genetic variant in pre-miR-27a is associated with a reduced renal cell cancer risk in a Chinese population. PLoS One 2012;7:e46566. PMID: 23118855.



20. Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci U S A 2007;104:16170–16175. PMID: 17911264.



21. Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, et al. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem 2010;285:17869–17879. PMID: 20371610.



22. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513–10518. PMID: 18663219.



23. El-Hefnawy T, Raja S, Kelly L, Bigbee WL, Kirkwood JM, Luketich JD, et al. Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin Chem 2004;50:564–573. PMID: 14718398.


24. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997–1006. PMID: 18766170.


25. Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem 2008;54:482–490. PMID: 18218722.


26. Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PLoS One 2008;3:e3148. PMID: 18773077.



27. Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer 2010;102:1174–1179. PMID: 20234369.



28. Liu CJ, Kao SY, Tu HF, Tsai MM, Chang KW, Lin SC. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis 2010;16:360–364. PMID: 20233326.


29. Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 2010;127:118–126. PMID: 19876917.


30. Yamamoto Y, Kosaka N, Tanaka M, Koizumi F, Kanai Y, Mizutani T, et al. MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers 2009;14:529–538. PMID: 19863192.


31. Wang Y, Lee CG. MicroRNA and cancer: focus on apoptosis. J Cell Mol Med 2009;13:12–23. PMID: 19175697.



32. Ferracin M, Lupini L, Salamon I, Saccenti E, Zanzi MV, Rocchi A, et al. Absolute quantification of cell-free microRNAs in cancer patients. Oncotarget 2015;6:14545–14555. PMID: 26036630.



33. Du M, Shi D, Yuan L, Li P, Chu H, Qin C, et al. Circulating miR-497 and miR-663b in plasma are potential novel biomarkers for bladder cancer. Sci Rep 2015;5:10437. PMID: 26014226.




34. Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res 2011;39:7223–7233. PMID: 21609964.



35. Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423–433. PMID: 21423178.



36. Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer 2011;47:784–791. PMID: 21112772.


37. Ma Y, Yu S, Zhao W, Lu Z, Chen J. miR-27a regulates the growth, colony formation and migration of pancreatic cancer cells by targeting Sprouty2. Cancer Lett 2010;298:150–158. PMID: 20638779.


38. Lerner M, Lundgren J, Akhoondi S, Jahn A, Ng HF, Akbari Moqadam F, et al. MiRNA-27a controls FBW7/hCDC4-dependent cyclin E degradation and cell cycle progression. Cell Cycle 2011;10:2172–2183. PMID: 21597324.


Fig. 1
Expression levels of three miRNAs in the screening cohort. miR-21 (A), miR-27a (B), and miR-155 (C). The expression level of each miRNA was normalized to U6snRNA.
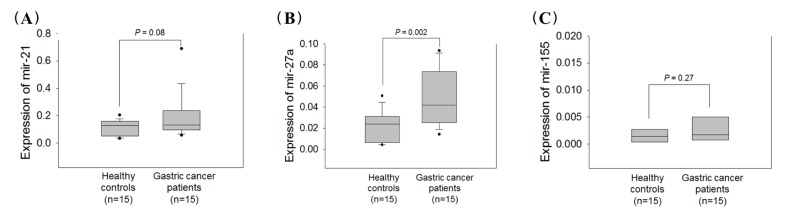
Fig. 2
miR-27a expression level in paired gastric cancer tissues. (A) Box plot of miR-27a expression level (n= 73). (B) MiR-27a expression level by TNM stage. (C) The receiver-operator characteristic curve and area under the receiver-operator characteristic curve (AUC) value were estimated using R software. The miR-27a expression level was normalized to U6snRNA. (D) Kaplan-Meier analysis of patient survival based on miR-27a expression in gastric tumors. The log-rank test was used to compare survival between the low- and high- miR-27a expression groups of patients on the basis of average miR-27a expression.
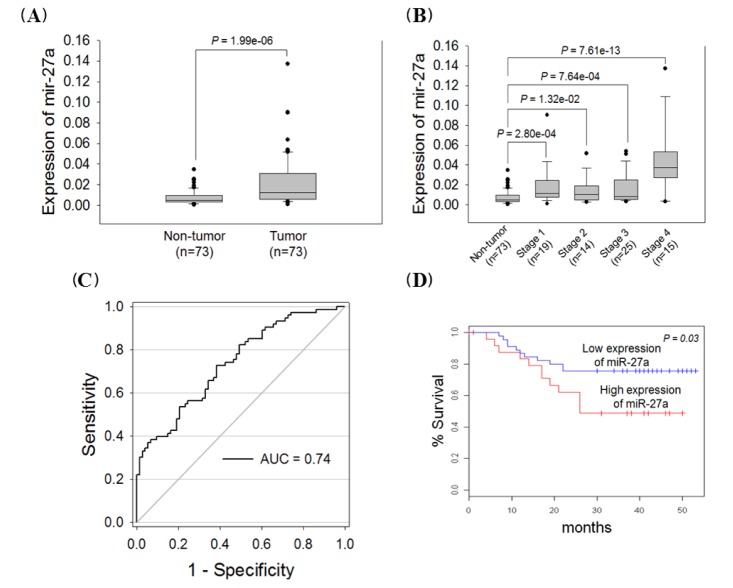
Fig. 3
Plasma miR-27a expression levels in the validation cohort. (A) Box plot of plasma miR-27a expression level. (B) MiR-27a expression by TNM stage. (C) Receiver-operator characteristic (ROC) curve analysis using the plasma miR-27a expression level. The plasma miR-27a marker yielded an area under the ROC curve (AUC) of 0.70 with 75% sensitivity and 56% specificity in discriminating gastric cancer.
- TOOLS
-
METRICS

-
- 16 Crossref
- 7,130 View
- 88 Download





