Examining the Gm18 and m1G Modification Positions in tRNA Sequences
Article information
Abstract
The tRNA structure contains conserved modifications that are responsible for its stability and are involved in the initiation and accuracy of the translation process. tRNA modification enzymes are prevalent in bacteria, archaea, and eukaryotes. tRNA Gm18 methyltransferase (TrmH) and tRNA m1G37 methyltransferase (TrmD) are prevalent and essential enzymes in bacterial populations. TrmH involves itself in methylation process at the 2'-OH group of ribose at the 18th position of guanosine (G) in tRNAs. TrmD methylates the G residue next to the anticodon in selected tRNA subsets. Initially, m1G37 modification was reported to take place on three conserved tRNA subsets (tRNAArg, tRNALeu, tRNAPro); later on, few archaea and eukaryotes organisms revealed that other tRNAs also have the m1G37 modification. The present study reveals Gm18, m1G37 modification, and positions of m1G that take place next to the anticodon in tRNA sequences. We selected extremophile organisms and attempted to retrieve the m1G and Gm18 modification bases in tRNA sequences. Results showed that the Gm18 modification G residue occurs in all tRNA subsets except three tRNAs (tRNAMet, tRNAPro, tRNAVal). Whereas the m1G37 modification base G is formed only on tRNAArg, tRNALeu, tRNAPro, and tRNAHis, the rest of the tRNAs contain adenine (A) next to the anticodon. Thus, we hypothesize that Gm18 modification and m1G modification occur irrespective of a G residue in tRNAs.
Introduction
Marinobacter species is one of the most ubiquitous classes formed in the marine ecosystem. It occurs throughout the water column in deep oceans, and it exerts a significant impact on various biogeochemical cycles. Marinobacter aquaeolei is involved in redox reactions that utilize oxygen and nitrate as terminal electron acceptors [1]. It exhibits hydrocarbon-degrading mechanisms and is capable of diverse extremophilic attributes (psychrophily, oligotrophy, and halotolerance) [2]. Marinomonas MWYL1 is able to grow on the betaine molecule and use dimethylsulfoniopropionate (DMSP) as their sole carbon source. Dimethyl sulphide is released as a by-product of metabolism of DMSP and reaches the atmosphere, which subsequently forms cloud condensation nuclei during oxidation [3]. The key gene (dddD) in M. MWYL1 encodes for acetyl-CoA transferase, which adds CoA to DMSP, prior to its subsequent cleavage [4]. Thus, M. MWYL1 is involved in the cycling of the climate-changing gas DMS, and their genomic potential still needs to be unraveled. Nitrobacter hamburgensis, a gram-negative bacterium, forms a biofilm, which is considered a suitable alternative for the treatment of wastewater that otherwise requires large and expensive reactors for efficient bioremediation [5]. Nitrobacter winogradskyi inhabits many soil types, natural stones, as well as both fresh and salt water. It derives its energy at once through nitrite oxidation and carbon dioxide fixation, thus acting as a chemolithoautotroph. The gammaproteobacterium class of Nitrosococcus oceani is an obligate chemolithoautotroph capable of extracting energy and reducing power from the oxidation of ammonia to nitrite. N. oceani's impact on the ecosystem is the massive release of nitrogen oxide and nitrogen gases into the atmosphere, thus completing the nitrification cycle. It also contributes to the maintenance of concentrations of nitrate in the nitrogen pool of the deep ocean at 40 µM [6].
Insight into tRNA modifications
tRNA is a small chain of nucleotides that decipher the genetic code to protein synthesis on codon and anticodon interactions and has a potential role in DNA repair mechanism tRNA methyltransferase brings changes in the nucleosides in the tRNA structure. Recently, some of these nucleoside modifications have been reported to be involved in the cell response to environmental stress and in the repair of DNA damage as well. In many cases, the posttran-scriptional modification of tRNA has facilitated translation of deviant sense codons [7], and these modifications are essential for tRNA folding function. The amino acyl tRNA synthetase recognizes the cognate tRNA through their structure and chemistry, contributed by modified nucleosides at the anticodon loop [8]. Thus, the modification of stem and loop structures of tRNAs regulates gene expression by ordering conformation and dynamics for recognition of the codon and maintenance of the reading frame. Similarly, mRNAs are programmed to shift their frame when there is no modification in tRNA [8]. In an actively dividing cell, tRNA gene content determines the relative abundance of tRNA isoacceptor, which, in turn, determines codon translation efficiency [9]. The relative abundance of a particular tRNA isoacceptor is kingdom-specific, and enzymes present in the organism increase the translational efficiency of codons through modification in the anticodon wobble base. It implies that the modified enzymes play a major role in the evolution of genomic composition [9]. Methylation reactions account for the majority of posttranscriptional modifications in tRNA [10]. tRNA methyltransferase (Trm) catalyzes the modification along the length of tRNA, including at the anticodon site. Indeed, Trm has a potential and crucial role in enhancing the synthesis of proteins that participate in the damage response [11].
tRNA Gm18 methyltransferase (TrmH)
To date, 150 modified nucleosides have been identified in RNA, and in tRNA alone 92 such modifications have been recorded [12]. Prominent among those are 2'-OH ribose methylated nucleoside, which is commonly found in tRNAs. The posttranscriptional modification in RNA is mediated through specific methyltransferase enzymes [13]. These modifications can occur in the noncoding region of the RNA sequences. In fact, tRNA contains abundant modified nucleosides, which stabilize the L-shaped tRNA structure and improves their molecular recognition [14]. A conserved guanosine is present at position 18 in the D loop of t-RNA modified to 2'-OH methyl guanosine. This modification stabilizes the L-shaped structure of t-RNA by interacting with pseudouridine 55 [15]. The enzyme TrmH catalyzes the reaction through transfer of a methyl group from S-adenosyl-methionine (AdoMet) to the 2'-OH group of the G18 ribose in tRNA D-loops [16]. The TrmH gene in Escherichia coli [17], Thermus thermophilus [18], and Aquifex aeolicus [19] has been identified to have similar catalytic motifs in them. TrmH is subdivided into two classes based on their substrate tRNA specificity. The type I enzymes are involved in the modification of all tRNA species, whereas type II modifies only a subset of tRNA species [20]. The Gm18 modification contributes towards defense reactions during pathogen encounters and has been observed in high frequency both in eukaryotes and prokaryotes, including viral RNA [21]. Therefore, a Gm18 modification study gains a significant factor of an infectious microbe, as it acts as a toll-like receptor 7 antagonist [21], and it can be utilized as an anti-inflammatory drug as well [20].
tRNA m1G37 methyltransferase (TrmD)
Lots of modified nucleosides are present at the region of anticodons, specifically at positions 34 (the wobble position) and 37 (3' and adjacent to the anticodon). One of them is the 1-methyl guanosine (m1G) modification occurring at position 37 in tRNA [22]. This modification is catalyzed by TrmD, which catalyzes the addition of a methyl group from AdoMet to G at position 37, adjacent to the anticodon [23]. The specificity of this enzyme is determined by V, T, and D side loops and the presence of a G36pG37 sequence [24]. TrmD is a vital enzyme for maintaining the correct reading frame during translation [25]. Lack of this modification at the 37th position (i.e., next to the anticodon) leads to a +1 frameshift and hinders the translation efficiency [26]. The nucleus-encoded tRNA is methylated at G before being transported to the cytoplasm, whereas in mitochondria, the tRNA is methylated at G37 by a nucleus-encoded enzyme and transported into organelles [27]. Both TrmH and TrmD enzymes are under the class of the SpoU family and share similar catalytic reactions [28]. Hence, the study confirms that tRNA subsets contain G bases at the 18th position (D-loop) and next to the anticodon (anticodon loop).
Methods
FASTA format of tRNA gene sequences of M. aquaeolei, M. MWYL1, N. hamburgensis, N. winogradskyi, and N. oceani were retrieved from a tRNA database (http://gtrnadb.ucsc.edu/) [29] and used for multiple sequence alignment by BioEdit software (http://www.mbio.ncsu.edu/bioedit/bioedit.html) [30].
Results
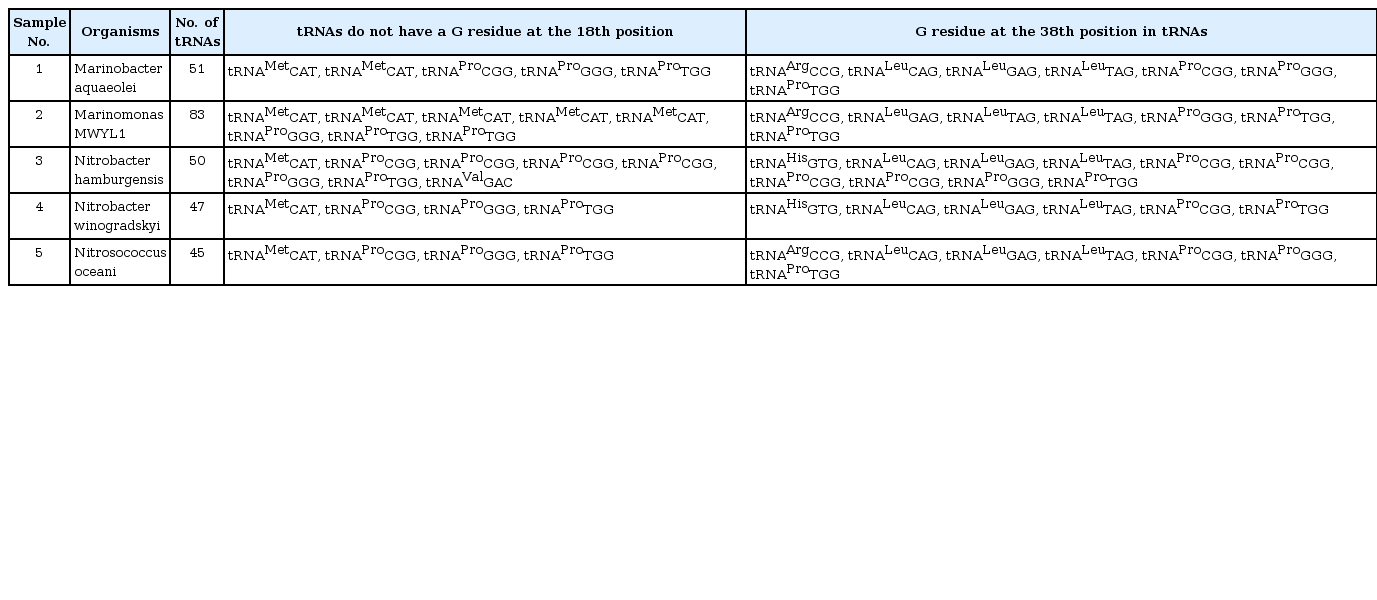
The presence of Gm18 modification bases were analyzed in total tRNAs of M. aquaeolei, M. MWYL1, N. hamburgensis, N. winogradskyi, and N. oceani. These organisms' tRNA contains a G residue at the 18th position adjacent to the anticodon loop, except tRNAMet, tRNAPro, and tRNAVal (Table 1). Furthermore, the 18th position of tRNAs in M. aquaeolei (51 tRNAs) and M. MWYL1 (83 tRNAs) contain thymine (T), cytosine (C), or adenine (A) instead of G residue. M. aquaeolei's tRNAMetCAT (two copies), tRNAProCGG, tRNAProCGG, and tRNAProTGG have T instead of G residues. M. MWYL1 contains 9 copies of tRNAMetCAT; of these, six tRNAs have a T residue at the 18th position and three contain a G residue at the 18th position. Only three proline-specific tRNAs are formed in M. MWYL1, and all of them show a T base at the 18th position instead of G. The organism N. hamburgensis comprises 50 tRNA subsets; among these, eight tRNAs showed no G residue at the 18th position, and the rest of the 42 had a G residue. Thus, the tRNAs without G residues are tRNAMetCAT, tRNAProCGG (three copies), tRNAProGGG, tRNAProTGG, and tRNAValGAC. Therefore, eight tRNAs at the 18th position show T, C, or A. Besides tRNAMetCAT, tRNAProCGG and tRNAProGGG show a C residue, and tRNAProCGG and tRNAProTGG hold a T residue at the 18th position. Nevertheless, tRNAValGAC confirmed an A residue at the 18th position. Among six proline-specific tRNAs (tRNAProCGG), only one showed a G residue at the 18th position. Furthermore, N. winogradskyi and N. oceani revealed 47 and 45 tRNAs, respectively. These two organisms hold similar properties-i.e., tRNA subsets, like tRNAMetCAT, tRNAProCGG, tRNAProGGG, and tRNAProTGG, containing a T residue at the 18th position as a substitute for the G residue.
Locating guanosine next to the anticodon in tRNAs
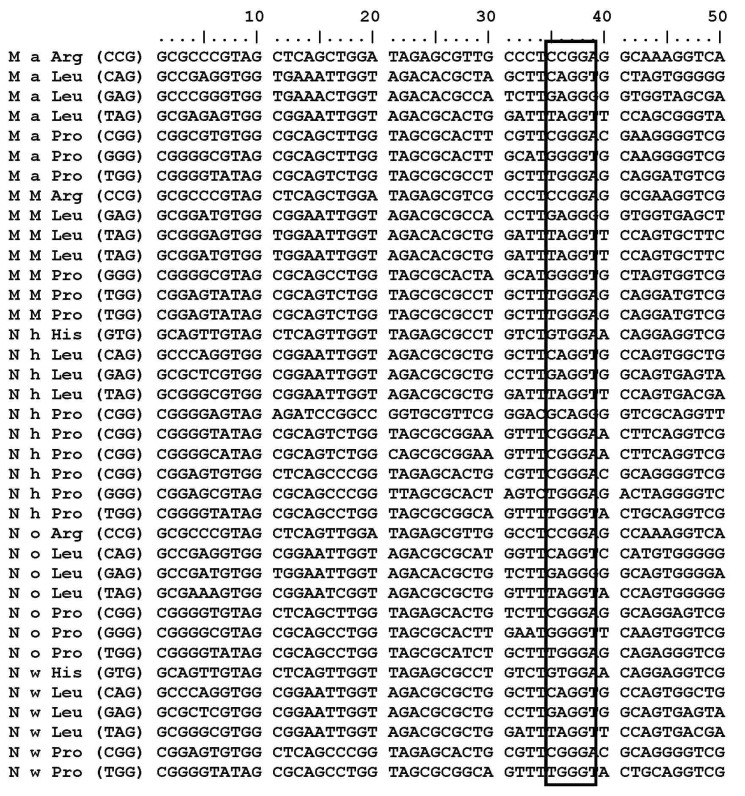
M. aquaeolei, M. MWYL, N. hamburgensis, N. winogradskyi, and N. oceani have common conserved tRNA subsets (tRNAArg, tRNALeu, and tRNAPro) that contain a G residue at the 38th position (G37pG38) next to the anticodon. Variations of the G residue in archaea Methanococcus jannaschii tRNAs, such as tRNACys (G35pG36); tRNAHis, and tRNAArg (G36pG37); tRNAGln and tRNATry (G37pG38); tRNAArg, tRNAPro, and tRNASer (G37pG39); tRNALeu, tRNAGlu, tRNATrp, tRNASer, and tRNAPro (G39pG40) [31] are already revealed. M. aquaeolei, M. MWYL1, and N. hamburgensis share similar tRNA subsets (tRNAArg, tRNALeu, and tRNAPro) that contain a G residue at the 38th position (G37pG38) next to the anticodon (Table 1). N. winogradskyi loses the G residue in tRNAArg, but it has presented it at the 38th position in tRNAHisGTG, tRNALeuCAG, tRNALeuGAG, tRNALeuTAG, tRNAProCGG, tRNAProGGG, and tRNAProTGG. N. oceani contains conserved tRNAs (tRNAArgCCG, tRNALeuCAG, tRNALeuGAG, tRNALeuTAG, tRNAProCGG, tRNAProGGG, tRNAProTGG) in which the G residue (G37pG38) is presented next to the anticodon (Table 1). An interesting finding is that tRNAMet and tRNAPro are the only two tRNA copies that do not have a G residue at 18th position in all of these organisms (Fig. 1). Additionally, the tRNAArg, tRNALeu, and tRNAHis share common properties, except tRNAPro. But, tRNAPro has a G residue at the 18th and 38th positions in these five organisms (Fig. 2).
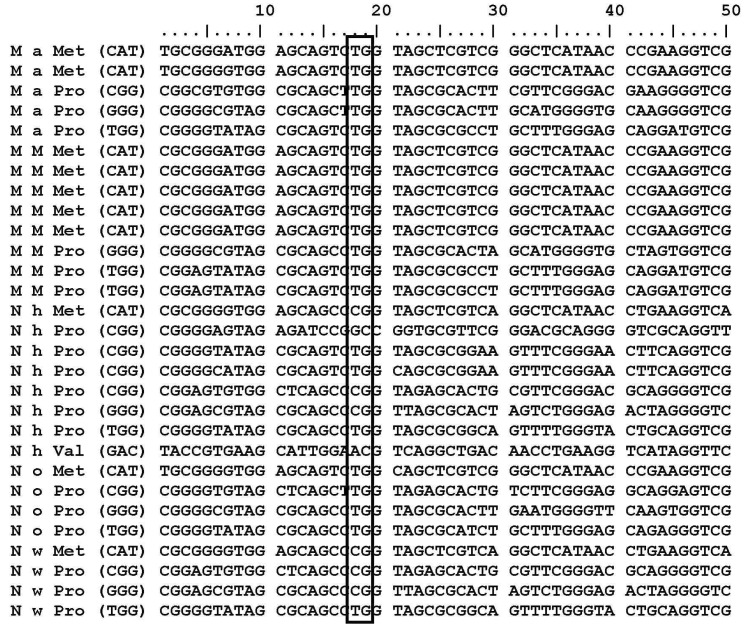
tRNAs sequences are marked at 18th positions, in all organism where tRNAMet and tRNAPro sequences hold A, T or C instead of G at 18th position.
Discussion
It is proposed that G18 modification base occurrence and non-occurrence in tRNA subsets in Marinobacter is attributed to the inhabitably extreme environments (M. aquaeolei, M. MWYL1), including chemolithotropic N. hamburgensis, N. winogradskyi, and N. oceani. Thus, a notable finding is that only methionine- and proline-specific tRNAs do not have a G residue at the 18th position, and even its absence at the 18th position does not hamper the possibility to methylate the 2'-OH group of ribose in tRNAs. It might be a modified Gm18 base (D loop) that interacts with a ψ55 tertiary base (T loop) for conformational rigidity of the tRNA structure [32]. Another finding is the m1G modification site in tRNAs, in which it was previously reported that only three types of tRNAs (tRNAArg, tRNALeu, and tRNAPro) held a G base next to the anticodon [22, 33]. Later, the m1G modification was reported in additional tRNAs, such as tRNAGln in archaea and tRNAAsp in eukaryotes [34]. A study reported tRNAGln with m1G in Mycobacterium tuberculosis H37Rv, tRNAPhe with m1G in Staphylococcus aureus MRSA252, and Streptococcus pneumoniae D39 [31]. The Gm18 modification base G is conserved in these five organisms. For instance, tRNAMet and tRNAPro sequences do not have a G residue at the 18th position. Furthermore, the N. hamburgensis organism does not have a G residue in tRNAValGAC. The m1G and its location are highly conserved and located at the 38th position in tRNAs. Mostly the purine bases G or A are placed next to the anticodon in all tRNAs. This modification may influence the codon-anticodon interaction in the translation process [35]. These findings suggest that either these two modifications are not necessary to occur in all tRNAs or might occur with/without G residues at the respective positions. Hence, we conclude that the Gm18 modifications and m1G37 tRNAs are essential in all three domains and have revealed a conserved tRNAs with m1G37 in a few marine and chemolithoautotrophs. Further study is called for to include more marine and extremophiles to gain insights into tRNA modification bases and their role in biological processes.