 |
 |
- Search
| Genomics Inform > Volume 20(2); 2022 > Article |
|
Abstract
The influenza A viruses have high mutation rates and cause a serious health problem worldwide. Therefore, this study focused on genome characterization of the viruses isolated from Thai patients based on the next-generation sequencing technology. The nasal swabs were collected from patients with influenza-like illness in Thailand during 2017-2018. Then, the influenza A viruses were detected by reverse transcription-quantitative polymerase chain reaction and isolated by MDCK cells. The viral genomes were amplified and sequenced by Illumina MiSeq platform. Whole genome sequences were used for characterization, phylogenetic construction, mutation analysis and nucleotide diversity of the viruses. The result revealed that 90 samples were positive for the viruses including 44 of A/H1N1 and 46 of A/H3N2. Among these, 43 samples were successfully isolated and then the viral genomes of 25 samples were completely amplified. Finally, 17 whole genomes of the viruses (A/H1N1, n=12 and A/H3N2, n=5) were successfully sequenced with an average of 232,578 mapped reads and 1,720 genome coverage per sample. Phylogenetic analysis demonstrated that the A/H1N1 viruses were distinguishable from the recommended vaccine strains. However, the A/H3N2 viruses from this study were closely related to the recommended vaccine strains. The nonsynonymous mutations were found in all genes of both viruses, especially in HA and NA genes. The nucleotide diversity analysis revealed negative selection in the PB1, PA, hemagglutinin (HA) and neuraminidase (NA) genes of the A/H1N1 viruses. High-throughput data in this study allow for genetic characterization of circulating influenza viruses which would be crucial for preparation against pandemic and epidemic outbreaks in the future.
Currently, influenza viruses are still a major cause of respiratory disease and can affect all age groups, resulting in a serious public health problem. The estimated infection rate of influenza viruses is approximately 5% to 15% of the population [1]. Furthermore, there are more than 500,000 deaths reported worldwide [2]. Seasonal influenza is caused by influenza A (A/H1N1 and A/H3N2 subtypes) and influenza B (B/Victoria and B/Yamagata lineages) viruses. However, the influenza A viruses cause more severity, and lead to more epidemics and pandemics due to the high mutation rates which result from antigenic drifts and antigenic shifts [3]. First, antigenic drift is caused by the accumulation of point mutations that change the properties of the viral hemagglutinin (HA) and neuraminidase (NA) surface proteins to avoid the host immune system. On the other hand, an antigenic shift is a genetic reassortment process when at least two strains of influenza A viruses have infected within the same cell [4]. During viral replication, the high rate of mutation is promoted by error-prone polymerase enzyme [5]. The mutation rates of the influenza A virus have been reported within a range of 2.0├Ś10ŌłÆ6 to 2.0├Ś10ŌłÆ4 mutations per site per round of genome replication [6-8]. Therefore, this evidence suggests that each replicated genome of influenza A carries an average of 2ŌĆō3 mutations per genome [9]. The virus has gradually adapted to its antigenic sites to avoid the host immune response and vaccination [10]. Due to the high mutation rates, the influenza vaccine was less effective (only 29% to 61%) against seasonal outbreaks during 2019-2020 [11].
In Thailand, influenza transmission occurs year-round with two annual peaks: a major peak in the rainy season and a minor peak in winter [12]. Previous studies have reported that influenza was a major cause of morbidity and mortality in Thailand and resulted in crucial economic costs annually. A study conducted during 2005ŌĆō2008, estimated an annual average of 36,400 influenza-associated hospitalizations and 300 deaths occurred, with significantly higher mortality rates in children and the elderly [13]. Furthermore, several studies examined the genetic variabilities within HA and NA genes of influenza A viruses based on Sanger sequencing [14-17]. Interestingly, whole genome sequencing (WGS) can be applied to characterize viral strains and provide the comprehensive information of the influenza genome for better understanding of the viral evolution and novel viral strains [18].
Nowadays, next-generation sequencing (NGS) has the advantages of massively parallel sequencing thus making it the ideal tool for characterization of the viral whole genome, viral reassortment and viral mutations [18,19]. Consequently, WGS of influenza viruses based on NGS technology can provide the information necessary to understand the characteristics of influenza viruses. This study aimed to investigate the viral genome and mutations of influenza A viruses circulating in Thailand from 2017 to 2018, and this approach can be further applied for preparation against pandemic and epidemic outbreaks in the future.
The study protocol was approved by the Institutional Review Board (IRB No. 337/57) and Institutional Biosafety Committee (MDCU-IBC No. 001/2018) from the Faculty of Medicine, Chulalongkorn University. Briefly, nasal swab samples from patients with influenza-like illness (ILI) were obtained from Bangpakok 9 International Hospital and Chum Phae Hospital from August 2017 to November 2018. The clinical samples were preserved in viral transport media consisting of HankŌĆÖs Balanced Salt Solution supplemented with 1% bovine serum albumin, amphotericin B (15 ┬Ąg/mL), penicillin G (100 U/mL), and streptomycin (50 ┬Ąg/mL). The nasal swab samples were screened for influenza virus infection using a one-step multiplex reverse transcription-quantitative polymerase chain reaction (RT-qPCR) as described previously [20,21]. Briefly, the assay was performed in a 10 ┬ĄL final volume, containing 1 ┬ĄL of RNA sample, 5 ┬ĄL of 2├Ś reaction mix, 0.2 ┬ĄL of SuperScript III RT/Platinum Taq Mix (Invitrogen, Carlsbad, CA, USA), an additional 0.1 mM of MgCl2, 0.25 ┬ĄM of each primer, and 0.125 ┬ĄM of each probe. The one-step multiplex RT-qPCR was performed on the StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using the following thermal cycling conditions: at 55┬░C for 30 min for reverse transcription, followed by 95┬░C for 10 min, continuing with 40 cycles of 95┬░C for 15 s and 60┬░C for 30 s.
Madin-Darby canine kidney (MDCK) cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in DulbeccoŌĆÖs modified Eagle Medium (DMEM) with high glucose (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA) and 1% (v/v) penicillin/streptomycin (Gibco) maintained under humidified 5% CO2 at 37┬░C [22].
MDCK cells were used for influenza virus isolation and propagation as described in the previous study [23]. Briefly, MDCK cells were seeded in 60 mm tissue culture dishes (SPL Life Science, Pocheon, Korea) at 5├Ś105cells per dish in DMEM medium without antibiotics. When the cells reached around 80% confluence, the media were removed and then washed by phosphate buffer saline (PBS) (Amresco, Solon, OH, USA). Positive influenza samples were used for virus isolation. Briefly, 500 ┬ĄL of a nasal swab from influenza-positive samples was mixed with 500 ┬ĄL of DMEM with high glucose (HyClone) and filtered through 0.22 ┬Ąm filter (Millipore, Billerica, MA, USA). The filtrate was immediately processed to influenza viral propagation. Three hundred microliters of each filtered influenza-positive sample were mixed with 200 ┬ĄL infection medium (DMEM-high glucose supplemented with 2 mM L-glutamine and 0.5 ┬Ąg/mL TPCK-trypsin). The mixture was added in each dish and then incubated in 5% CO2 at 37┬░C for 1 h. After incubation, the virus suspension was removed and then washed with PBS. Finally, the cells were overlaid with fresh infection medium and incubated under 5% CO2 at 37┬░C for 48 h. After that, the cytopathic effect of infected cells was observed and the viral supernatant was collected. Each sample was isolated in three serial passages (P0ŌĆÆP2). The viral titers were quantified by RT-qPCR [24] .
One hundred and fifty microliters of the supernatant in each isolation passage were extracted using a GenUp Viral RNA kit (Biotechrabbit, Berlin, Germany) according to the manufacturerŌĆÖs instructions, and eluted in 60 ┬ĄL with warm RNase-free water. The concentration of total viral RNA was quantified by using Nanodrop UV spectrophotometer (Implen, Munchen, Germany). Three hundred nanograms per microliter of viral RNA were reverse transcribed into cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturerŌĆÖs instructions with 10 ┬ĄM reverse transcription primer (5╩╣-ACGCGTGATCAGCAAAAGCAGG-3╩╣) that is conserved and complemented with 12 nucleotides at the 3╩╣ ends of each influenza A viral genes [25]. The mixtures were incubated at 42┬░C for 1.5 h and heat-inactivated at 70┬░C for 10 min. The cDNAs were stored in ŌĆÆ20┬░C for further analysis.
To determine the amount of influenza virus in each sample passage, the viral matrix (M) gene was amplified based on StepOnePlus Real-time PCR Systems (Applied Biosystems) using SYBR Green Luna Universal qPCR Master Mix (New England Biolabs, Ipswhich, MA, USA) as described above. The results were analyzed by StepOnePlus Software v2.3 (Thermo Fisher Scientific). The samples amplified with Ct values lower than 28 were interpreted as positive influenza viral propagation [26].
The viral cDNAs from the previous step were used as templates for genome amplifications. The primer sets; forward primer (5╩╣-ACGCGTGATCAGCAAAAGCAGG-3╩╣) and reverse primer (5╩╣-ACGCGTGATCAGTAGAAACAAGG-3╩╣) were used for amplification of influenza A viral genes (8 segments) following the previous study [25]. Briefly, PCR master mix is composed of 1.25 ┬ĄM of each primer, 0.35 mM of dNTPs, 0.02 U/┬ĄL of Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific), 7.5 ┬ĄL of cDNA, and nuclease-free water to a final volume of 50 ┬ĄL. Subsequently, 15 ┬ĄL of the amplicons were analyzed by 1% agarose gel electrophoresis. The amplicons were purified by the QIAquick PCR Purification kit (Qiagen, Hilden, Germany) following the manufacturerŌĆÖs protocol. The concentrations of purified PCR products were measured by the Qubit dsDNA High-Sensitivity assay kit (Invitrogen).
The purified amplicons (1 ┬Ąg in 130 ┬ĄL) from the genome amplification step were sheared to approximately 200 bp fragments by the Covaris M220 Focused-ultrasonicator (Covaris, UK) with 20% duty factor, 50 unit of peak incident power (W), 200 cycles per burst for 150 s. The fragmented DNAs were used for DNA library preparation by using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs) following the manufacturerŌĆÖs instructions. Briefly, 50 ┬ĄL of DNA fragments were ends repaired and subsequently adapters ligated by using the NEB ligase master mix. Then, DNA libraries (approximately 320 bp) were cleaned up and size selected by AMPure XP beads (Beckman Coulter, Brea, CA, USA). For library enrichment, PCR amplification was carried out by adding the Illumina MiSeq-compatible indexes to the DNA libraries. Afterwards, the enriched DNA libraries were purified by 2% agarose gel electrophoresis with 100 V for 20 min and size selected (approximately 320 bp). Finally, the total DNA libraries were quantified by real-time PCR using the KAPA Library Quantification Kits (Kapa Biosystems, Wilmington, MA, USA). After that, the concentration of each sample was determined and pooled equally at 2 nM of each library. Subsequently, the pooled library was then diluted to 6 pM and paired-end sequenced (2├Ś150 bp) on an Illumina MiSeq instrument using MiSeq Reagent Kits v2 (300 cycles) according to the manufacturer protocol (Illumina).
The MiSeq Reporter Software version 2.4 was used for the primary analysis of FASTQ sequencing data. Low-quality reads (Q-score <30) and adaptors were trimmed. The passing filtered reads (Q-score Ōēź30) were aligned with the vaccine strains of influenza A reference genomes (A/California/07/2009 (H1N1) or A/South Australia/55/2014 (H3N2)) for genome characterization and mutation analysis by using CLC Genomics Workbench software (Qiagen). Mutation patterns and frequencies were generated by using GraphPad Prism version 6.01 software (GraphPad Software Inc., San Diego, CA, USA). The FASTQ files and FASTA files of influenza genome sequences were submitted to the Sequence Read Archive (BioProject ID: PRJNA576776) and GenBank as shown in Supplementary Table 1.
In this study, the HA and NA deduced amino acid sequences of A/H1N1 and A/H3N2 were aligned with reference strains retrieved from the Global Initiative on Sharing All Influenza Data (GISAID) EpiFlu database by using the Clustal W program, implemented in the BioEdit sequence alignment editor software v.7.2.5 [27]. Phylogenetic analysis was performed by mean of the maximum likelihood method (1,000 bootstrapping replicates) and LG with Freqs.(+F) model (discreate gamma distribution with 5-rate categories and complete deletion data subset) using the MEGA X software [28].
The nucleotide diversity (ŽĆ) within each gene of influenza A/H1N1 and A/H3N2 viruses was evaluated by PoPoolation v.1.2.2 to investigate the genetic variations of viruses within the sample [29]. The sliding window analysis of nonsynonymous nucleotide variation (ŽĆN) was performed based on Syn-nonsyn-sliding.pl script with the window size of nine codons and a step size of one codon. The average corresponding ŽĆN values were calculated and plotted to a middle position of the windows to demonstrate the degree of nonsynonymous substitutions within eight viral gene segments. In addition, the nonsynonymous nucleotide variations (ŽĆN) per synonymous nucleotide variation (ŽĆS) were analyzed by the Syn-nonsyn-at-position.pl script to investigate the neutrality of selection in each segment. The ŽĆN/ŽĆS ratios per gene in each influenza subtype were calculated as the average value from individual samples. Lastly, a paired Wilcoxon signed-rank test (p < 0.05) was used to compare pooled average ŽĆN and ŽĆS values within each subtype of influenza viruses.
In this study, 500 nasal swab samples were collected from patients with ILI and detected for influenza A virus by RT-qPCR. Ninety samples were influenza A virus-positive samples including 48.9% (44 samples) of A/H1N1 and 51.1% (46 samples) of A/H3N2 as shown in Table 1. Among these 90 samples, 43 samples (29 of A/H1N1 and 14 of A/H3N2) were successfully isolated in the second passage (P2) of MDCK cells with Ct value ranging from 13 to 28 (Table 2).
From 43 isolated samples, 25 samples were completely amplified as a full genome including 17 samples of A/H1N1 and eight samples of A/H3N2. Finally, 17 samples (12 samples of A/H1N1 and five samples of A/H3N2) passed the quality control of libraries preparation for NGS as shown in Table 1. The result revealed that 17 whole genomes of influenza A viruses were successfully sequenced with an average of 424,151 total reads per sample, 232,578 mapped reads per sample, and 1,720 genome coverage per sample (Table 2). Therefore, these results were highly confident for genome annotation and mutation analysis. The FASTQ data were deposited as BioProject accession no. PRJNA576776 and influenza genome sequences were submitted into GenBank database as summarized in Supplementary Table 1.
The HA and NA deduced amino acid sequences were used for phylogenetic analysis. The sequences were aligned with the HA and NA deduced amino acid sequences of the influenza vaccine strains (southern hemisphere influenza seasons during 2012ŌĆÆ2019) recommended by the World Health Organization (WHO). The influenza A/H1N1 viruses isolated from this study during 2017-2018 belonged to genetic subclade 6B.1. Interestingly, the results demonstrated that A/H1N1 viruses were closely related to influenza (A/California/7/2009) strain and distinguishable from the recommended influenza vaccine strains for use in 2017-2019 (A/Michigan/45/2015 (H1N1)) as shown in Fig. 1A and 1B. On the other hand, the circulating A/H3N2 strains, classified into subclade 3C.2a1 and 3C.2a2, were comparatively more closely related to the 2017ŌĆÆ2018 WHO influenza vaccine strains (A/Hong Kong/4801/2014 (H3N2) and A/Singapore/INFIMH-16-0019/2016 (H3N2)) as shown in Fig. 2A and 2B.
The variations of nonsynonymous within influenza A viruses among the samples in this study are summarized in Fig. 3. As shown in Fig. 3A, strong signals appeared in the polymerase (PB2, PB1, and PA) genes as well as in the NP gene in A/H1N1 virus. However, the NA, M, and NS genes showed the low nonsynonymous variations. Interestingly, the HA gene contained the pattern of the variation signals around the middle position of this A/H1N1 gene. As for the results of A/H3N2 (Fig. 3B), the polymerase genes were presented as sharp and multiple peaks of the nonsynonymous nucleotide diversity. In addition, the HA, NP, and NA genes of A/H3N2 displayed sharp signals at the beginning and the end of these genes. Furthermore, the M and NS genes of the A/H3N2 only had peaks around the middle of the genes.
Exploring deeper detail about the direction of diversity, the ratios of nonsynonymous to synonymous nucleotide diversity (ŽĆN/ŽĆS) were introduced to examine the changes in nucleotide variations. In brief, the ŽĆN/ŽĆS ratios > 1 indicate that selective pressure promotes the new variations (positive selection). In contrast, the ŽĆN/ŽĆS ratios < 1 refer to the new variation being unfavored (negative selection). In addition, the ŽĆN/ŽĆS ratios Ōēł 1 suggest that neutral evolution occurs in these new variations. According to the results shown in Fig. 4, there was no significant positive selection occurring in this study. However, the statistically significant negative selections (p < 0.05) were found in PB1, PA, HA, and NA genes of A/H1N1. Meanwhile, the A/H3N2 exhibited random selections within these 10 genes due to there being no significant difference observed in the ŽĆN/ŽĆS ratios.
In this study, 90 out of 500 (18%) nasal swabs obtained from Thai patients with influenza-like-illness during 2017ŌĆÆ2018 were positive for influenza A virus detection based on RT-qPCR detection. The percentage of influenza A virus positive in this research was slightly higher than those reported in the previous study (13.2%) during 2016ŌĆÆ2017 in Thailand [30]. Previously, several studies have demonstrated that the appropriate quality and quantity of DNA are important for the successful NGS platform sequencing [31-33]. In particular, this study has successfully isolated 47.78% (43 of 90 samples) which are positively identified as the influenza A virus, which is higher than the previous study (3.04% of isolation rate) [34]. Also, the positive virus isolations (58.14%, 25 of 43 isolates) can be amplified with universal primers, following the study of Meinel et al. [35], which is appropriate for whole genome characterization and mutation analysis of influenza A virus. For NGS analysis, the result revealed that 17 whole genomes of influenza viruses were successfully sequenced with 232,578 mapped reads (424,151 total reads), average read length of 96.3 bp and average 1,720.4 genome coverage. Furthermore, the complete sequences of the viral genomes provide reliable and highly informative data despite the average genome coverages, depth coverage, which ranged from 237.7 to 4,229.7 (Table 2). The advantages of the NGS-based technique are that it provides the full genome segment and whole genome of influenza virus, as well as effectively reducing both the turnaround time and cost per nucleotide sequence for the whole genome when compared to the Sanger sequencing method [36-38]. However, the sequencing with the Sanger method does not provide the data for quasispecies and nucleotide diversity analysis. Interestingly, the NGS provides more information for minor mutations and selection pressures within the viral genome. Indeed, the nucleotide variations obtained from NGS can be applied for calculation of viral nucleotide diversity within each sample [39,40].
The number of mutations in the HA and NA genes of A/H1N1 might affect the efficiency of a vaccine, and related to deduced amino acid sequences of phylogenetic tree (Fig. 1A and 1B). The vaccine effectiveness of the 2017ŌĆÆ2018 flu vaccine against both influenza A viruses is approximately 25% to 52% in Europe and 27% to 44% in the United States [41,42]. Interestingly, the result of the influenza A/H1N1 phylogenetic tree with deduced amino acid sequences, which belongs to clade 6B.1, showed a long distance between vaccine strains for 2017ŌĆÆ2018 (A/Michigan/42/2015) and our A/H1N1 samples. This result implied that the vaccine might be less effective against A/H1N1 in Thailand. Moreover, the report from the US Centers for Disease Control and Prevention (CDC) also showed the vaccine effectiveness against A/H1N1 was 65% [43]. However, the phylogenetic analysis of both HA and NA deduced amino acid sequences revealed the closer relationship between A/H3N2 isolates (clade 3C.2a1 and 3C.2a2) and A/Hong Kong/4801/2014 strain which was the recommended vaccine virus for A/H3N2 [44]. Therefore, these results implied that the recommended vaccine was more effective against the influenza A/H3N2 in Thailand during 2017ŌĆÆ2018. Indeed, the phylogenetic trees of influenza A/H1N1 and A/H3N2 obtained in this study were correlated with recent genetic and antigenic characterizations of influenza viruses in Thailand [45].
In this study, the genome of circulating influenza A viruses in Thailand during 2017ŌĆÆ2018 was characterized. The result from NGS analysis not only provided the full genome of the virus but also acquired the amino acid substitutions across eight segmented genes. Moreover, there were several known functional mutations of influenza A/H1N1 that had been already characterized. Firstly, the mutations at I354, V344M, and S453T in the PB2 (Fig. 5A) could regulate in the cap-snatching from host RNAs during the viral RNA transcription process [46]. Furthermore, N321K in the PA was reported to increase the polymerase complex activity and the viral replication in the cell culture [46,47]. The amino acid substitution at V100I in the PA-X could trigger down-regulated innate immune response genes (Fig. 5C). Indeed, the amino substitutions at S91R, S181T, I312V, and E391K in the HA might be related to adaptive genetic variations that alter the salt bridge pattern and the membrane fusion stability for major antigenic sites and glycan specificity [46,47]. Three mutations (K180Q, S202T, and S220T) were located in the HA antigenic sites, which might be involved in the pathogenicity and contributed to the epidemic [48]. Moreover, the mutation (S220T) was observed to affect the infectivity and transmissibility of the virus in humans [49]. The mutation (R240Q) was found in the receptor-binding domain of the HA, which has been reported to increase virus growth [50]. The amino acid variations (D114N, K180Q, S202T, S220T, and K300E) were responsible for loss of antibody neutralization and decreased overall vaccine effectiveness (Fig. 5D) [51-53]. The amino acid substitutions (N44S, V241I, and N369K) in the NA have been reported to facilitate the stability of the virus [54]. The I188T and N449D mutations in the NA found in this study are similar to those reported in the previous study [55]; however, the function of the mutations has not been well characterized (Fig. 5F). Additionally, the nonsynonymous mutation at E55K, L90I, I123V, E125D, K131E, and N205S in the NS1 involves the inhibition of host gene expressions related to the interferon response [56,57]. Indeed, the E125D mutation in NS1 (Fig. 5H) interacts with cellular cleavage and polyadenylation specificity factor 30 (CPSF30), which is considered potential in host adaptation to influenza A/H1N1 virus [58,59].
In the influenza A/H3N2, the previous reports found R277Q and D69N at the antigenic epitope C, N137K, and N187K at the antigenic epitope D and E78K/G at the antigenic epitope E of the HA [60,61]. Among these, four amino acid substitutions (N137K, N187K, I422V, and G500E) belong to clade 3C.2a.1 and are represented by A/Singapore/INFIMH-16-0019/2016(H3N2) virus [62]. The T151N substitution in HA protein was related to the potential N-glycosylation site, affecting antigenic and other viral properties. Moreover, the Q327H substitution in the HA was suggested to bind host proteins (Fig. 6D) [62]. Since 2016, the accumulation of mutation at S245N of the NA has contributed to an N-glycosylation site. These mutations (S245N, S247T, and P468H) were introduced to the NA antigenic drift of the circulating A/H3N2 virus [63]. However, N329S mutation could result in a loss of N-glycosylation in the NA [64]. The V303I substitution has been observed in the NA protein (Fig. 6F) with a low resistance to NA inhibitors [65]. Indeed, most mutations of influenza A viruses observed in this study were identified as novel mutations which have not been reported yet (Figs. 5 and 6). However, the function of the novel mutations needs to be further investigated.
Nonsynonymous (ŽĆN) and synonymous (ŽĆS) mutations of the viruses can be accessed by NGS leading to nucleotide diversity (ŽĆN/ŽĆS) analysis. According to the previous study, the deep sequencing of A/Wisconsin/67/2005 (H3N2) revealed that the positive selection was observed in the viruses isolated from the chicken kidney, Vero cell culture, and embryonated chicken eggs, whereas the negative selection was found in virus from direct intranasal inoculation in the human challenge [40]. There was no significant nucleotide diversity observed in A/H3N2 viruses in our study, and this might be due to the strain of the virus, host cell, or limited numbers of the sample. For ŽĆN/ŽĆS analysis of influenza A/H1N1 viruses, the mutations existing in the viral genes with statistical significance were PB1, PA, HA, and NA genes in which these mutations were suggested as negative selection. Therefore, to further investigate the ŽĆN and ŽĆS variations, the sliding window analysis of those genes was performed to ensure that the negative selections were not the outcome of the averaging value across the entire gene. The results of sliding window analysis were consistent with the negative selections from the ŽĆN/ŽĆS analysis in those genes at which the ŽĆN signals were high and sharp at some regions of the genes, while the rest of the genes were relatively low in the ŽĆN signals.
In summary, the NGS was successfully applied for whole genome characterizations of influenza A/H1N1 and A/H3N2 viruses that provide the high-throughput data for phylogenetic construction, mutation analysis, and nucleotide diversity. The results revealed that the recommended vaccine A/H1N1 strain might be less effective against the A/H1N1 virus. Moreover, several mutations were demonstrated in both A/H1N1 and A/H3N2, especially in HA and NA genes. Finally, the negative selections were found in the PB1, PA, HA, and NA genes of the A/H1N1. Unfortunately, limited number of samples were successfully propagated, amplified, and sequenced in this study. Nevertheless, the whole genome data obtained from this study might be useful for mutation analysis and can be compared with data obtained from other studies in the future.
Notes
Acknowledgments
This work was partly supported by a grant from the National Science and Technology Development Agency (NSTDA) (P-17-51377), the Royal Golden Jubilee (RGJ) Ph.D. Programme scholarship (PHD/0150/2558), National Research Council of Thailand (NRCT)[2564NRCT321520] and Thailand Research Fund (TRF), the Chulalongkorn University Center of Excellence in Systems Biology and Chulalongkorn Academic Advancement into its 2nd Century Project. We would like to express gratitude to all the members of the Center of Excellence in Clinical Virology (Miss Preeyaporn Vichaiwattana and Mr. Sumeth Korkong), Faculty of Medicine, Chulalongkorn University for facilitating the sample collection. Moreover, we would like to thank the Department of Virology, Faculty of Veterinary Medicine Chulalongkorn University for support in influenza virus isolation (Assistant Prof. Aunyaratana Thontiravong, D.V.M., Ph.D.).
Supplementary Materials
Supplementary data can be found with this article online at http://www.genominfo.org.
Supplementary┬ĀTable┬Ā1.
The summary of accession numbers of influenza A virus in this study
Fig.┬Ā1.
The phylogenetic analysis of influenza A viruses (H1N1) circulating in Thailand during 2017ŌĆÆ2018 (diamond) compared with several World Health Organization recommended influenza vaccine strains (black triangle). The hemagglutinin (HA) (A) and neuraminidase (NA) (B) deduced amino acid sequences were analyzed based on mean of maximum likelihood with 1,000 bootstrapping and LG with Freqs. (+F) model (discreate gamma distribution with 5-rate categories and complete deletion data subset).
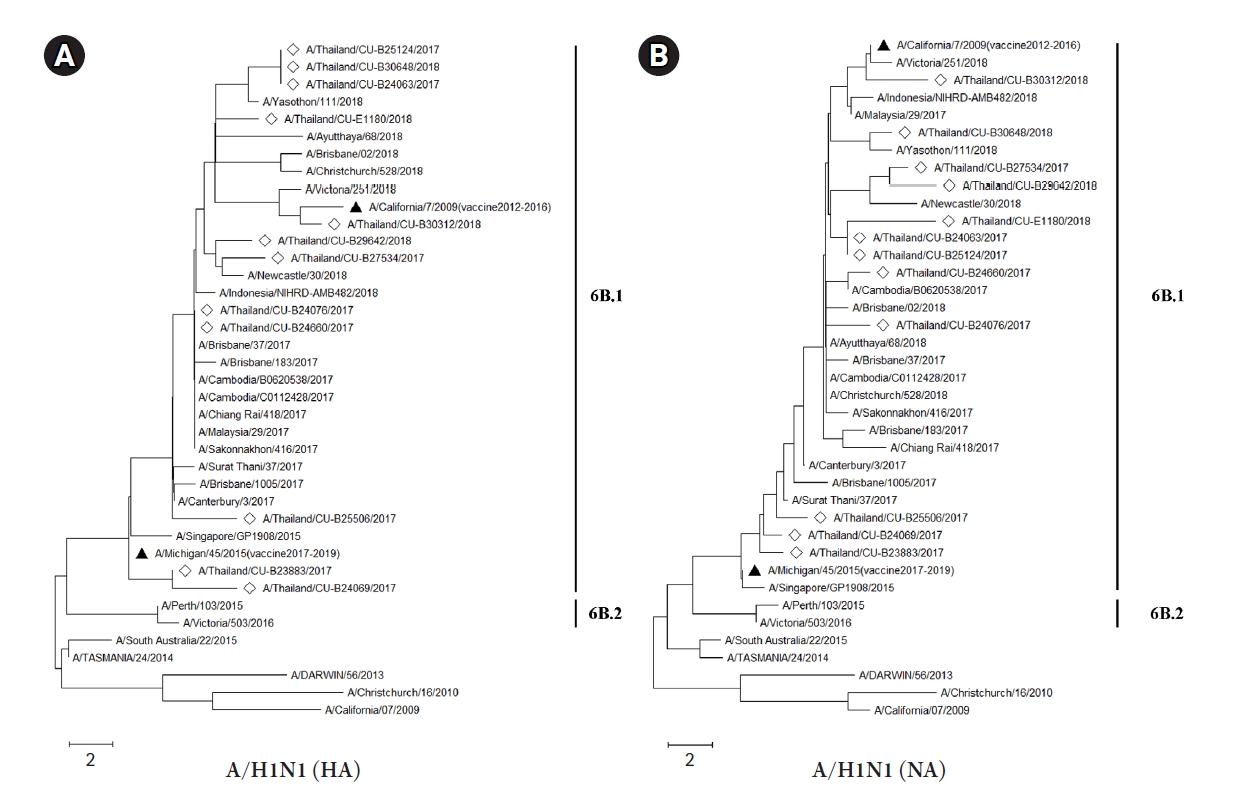
Fig.┬Ā2.
The phylogenetic analysis of influenza A viruses (H3N2) circulating in Thailand during 2017ŌĆÆ2018 (diamond) compared with several WHO recommended influenza vaccine strains (black triangle). The hemagglutinin (HA) (A) and neuraminidase (NA) (B) deduced amino acid sequences were analyzed based on maximum likelihood with 1,000 bootstrapping and LG with Freqs. (+F) model (discreate gamma distribution with 5-rate categories and complete deletion data subset).
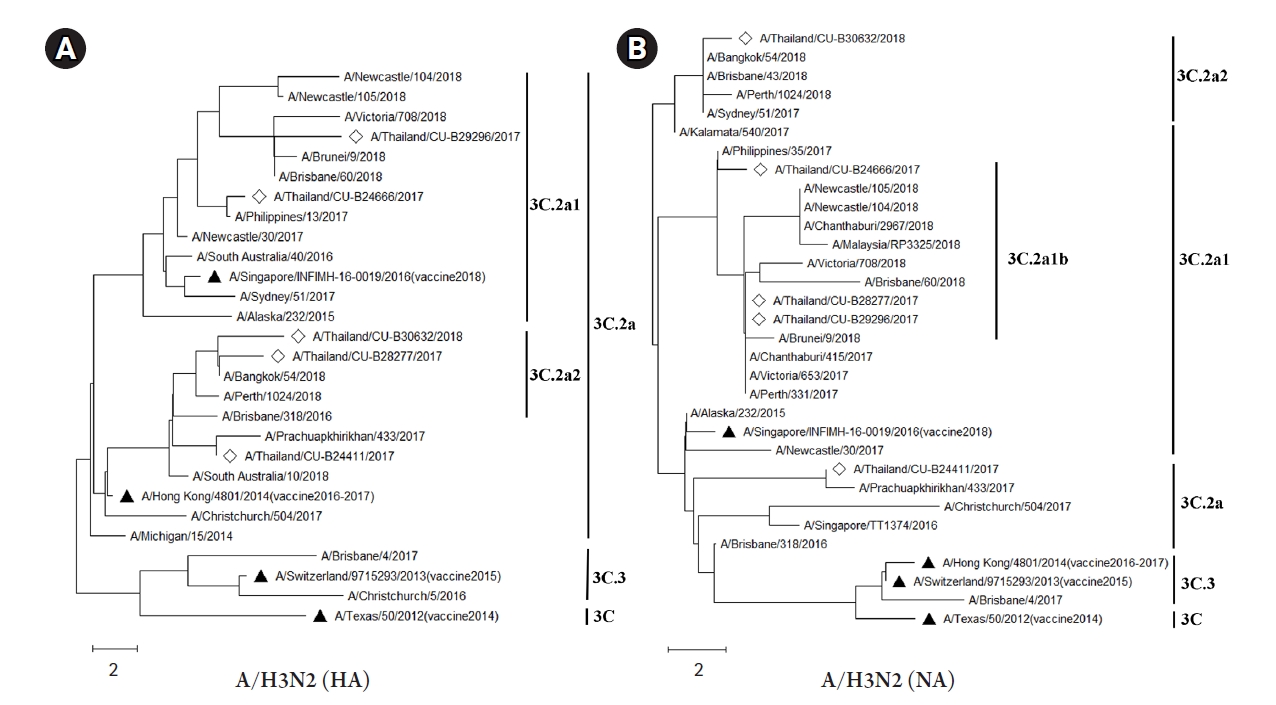
Fig.┬Ā3.
Sliding windows analysis of nonsynonymous nucleotide variation (ŽĆN) in eight genes of influenza A virus subtypes H1N1 (A) and H3N2 (B). The ŽĆN values were determined by sliding windows with the window size of nine codons and a step size of one codon. The mean corresponding ŽĆN values were calculated and plotted to a middle site of the windows.
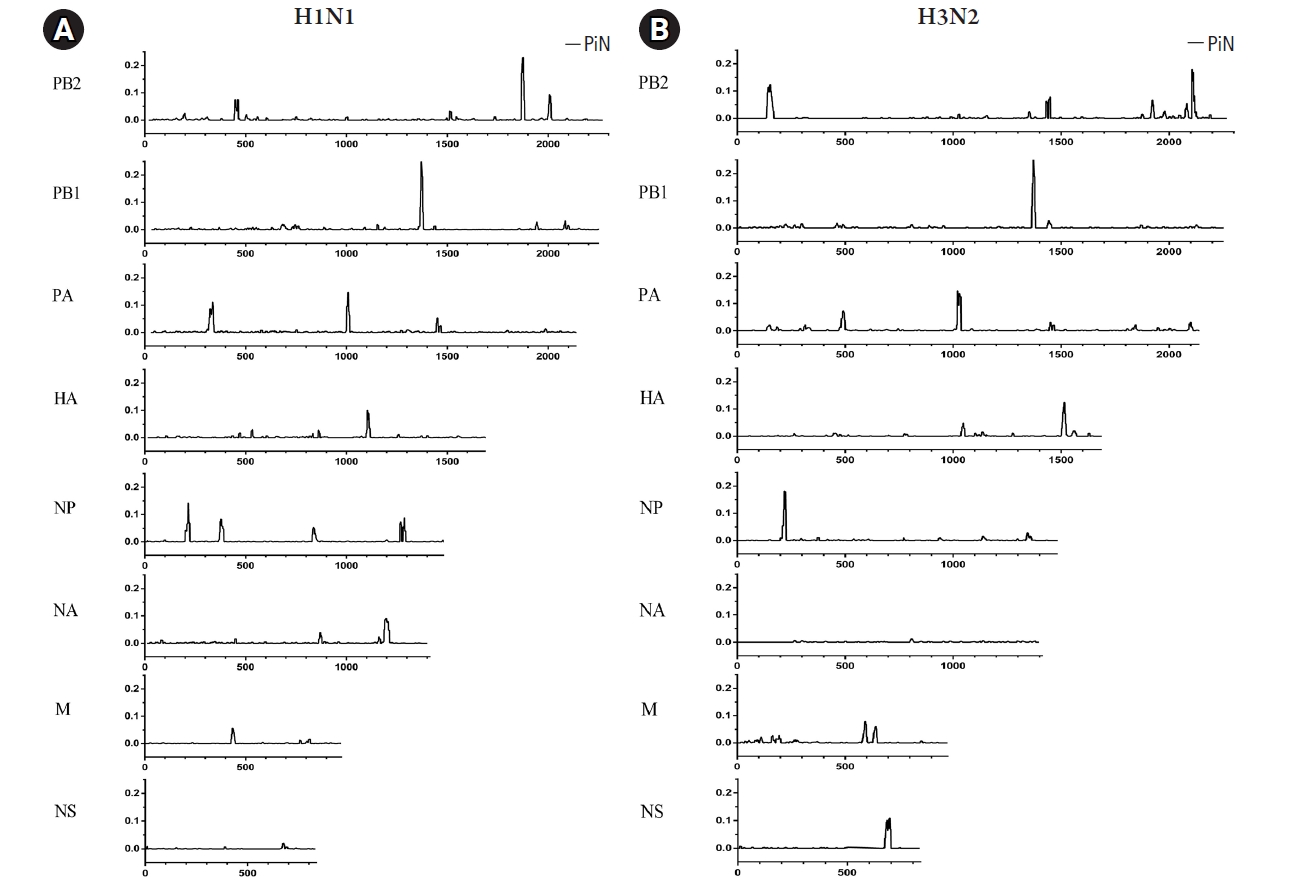
Fig.┬Ā4.
The ratio of nonsynonymous nucleotide variation (ŽĆN) to synonymous nucleotide variation (ŽĆS) analysis in eight genes of influenza A virus subtypes H1N1 (A) and H3N2 (B). Significant at *p < 0.05, **p < 0.01 and ***p < 0.001. The ratio ŽĆN/ŽĆS > 1: positive selection; ŽĆN/ŽĆS < 1: negative selection; ŽĆN/ŽĆS Ōēł 1: neutral evolution. The mean ŽĆN/ŽĆS, standard deviation (S.D.), and p-value (StudentŌĆÖs t-test) in each segment were summarized at the bottom of the figure.
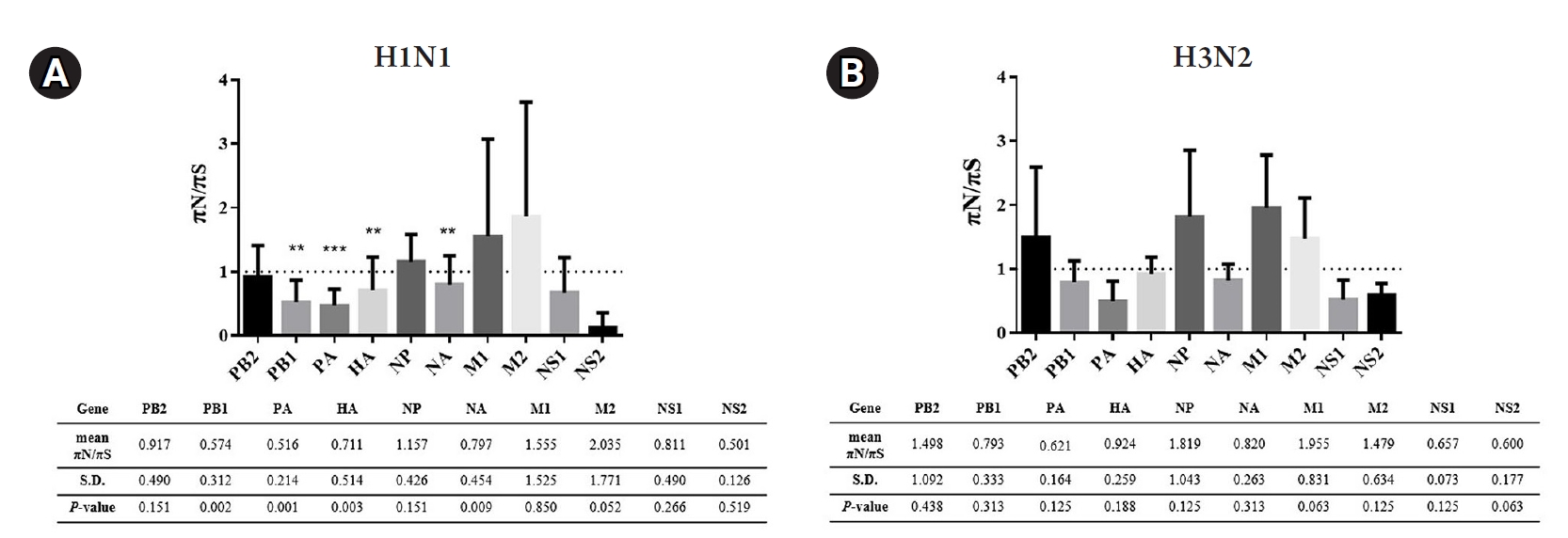
Fig.┬Ā5.
Mutation patterns with actual mutations frequencies observed in each viral gene segments of influenza A virus (H1N1): (A) PB2, (B) PB1, (C) PA, (D) HA, (E) NP, (F) NA, (G) M, and (H) NS. Amino acid changes were compared to the reference sequence (A/California/07/2009 (H1N1)).
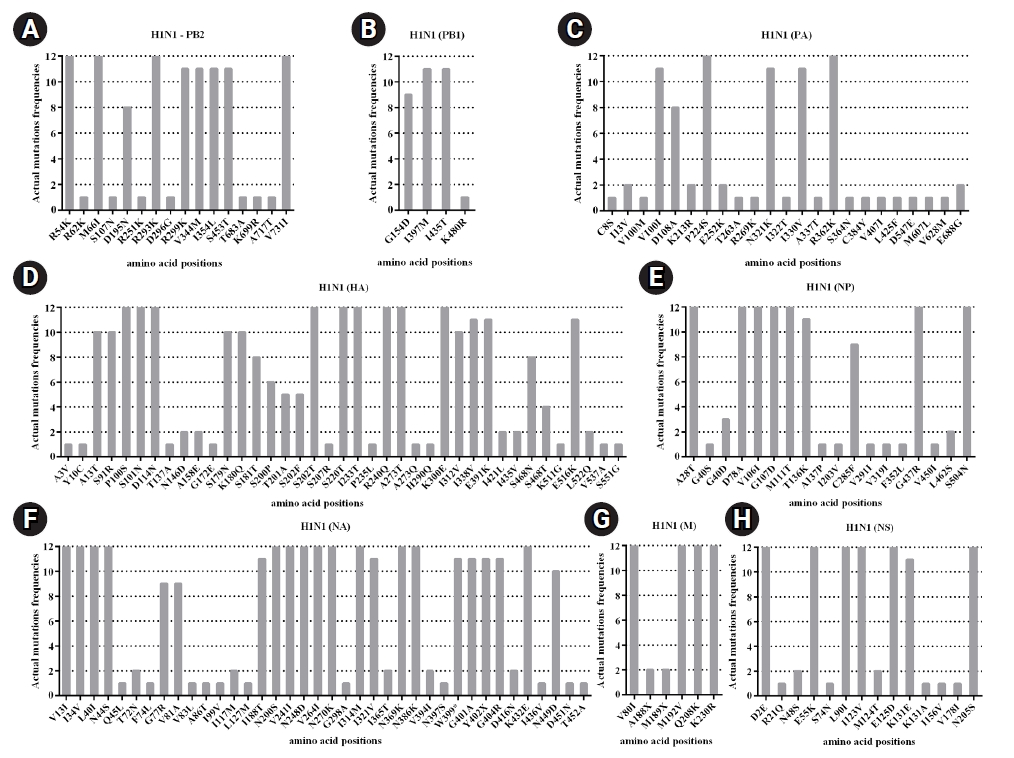
Fig.┬Ā6.
Mutation patterns with actual mutations frequencies observed in each viral gene segments of influenza A virus (H3N2). Amino acid changes were compared to the reference sequence (A/South Australia/55/2014 (H3N2)).

Table┬Ā1.
The amount of positive influenza A samples obtained from RT-qPCR, virus isolation, genome amplification, and NGS
| Positive samples | Positive virus isolation | Genome amplification | NGS | |
|---|---|---|---|---|
| Influenza A/H1N1 | 44 | 29 | 17 | 12 |
| Influenza A/H3N2 | 46 | 14 | 8 | 5 |
| Total | 90 | 43 | 25 | 17 |
Table┬Ā2.
The sample characteristic, virus isolation and NGS data of influenza A virus in this study
References
2. World Health Organization. Up to 650000 people die of respiratory diseases linked to seasonal flu each year. Geneva: World Health Organization, 2017. Accessed 2021 Dec 10. Available from: http://www.who.int/mediacentre/news/releases/2017/seasonal-flu/en/.

3. Shao W, Li X, Goraya MU, Wang S, Chen JL. Evolution of influenza A virus by mutation and re-assortment. Int J Mol Sci 2017;18:1650.



4. Cox NJ, Subbarao K. Global epidemiology of influenza: past and present. Annu Rev Med 2000;51:407ŌĆō421.


5. Boivin S, Cusack S, Ruigrok RW, Hart DJ. Influenza A virus polymerase: structural insights into replication and host adaptation mechanisms. J Biol Chem 2010;285:28411ŌĆō28417.



6. Bloom JD. An experimentally determined evolutionary model dramatically improves phylogenetic fit. Mol Biol Evol 2014;31:1956ŌĆō1978.



7. Nobusawa E, Sato K. Comparison of the mutation rates of human influenza A and B viruses. J Virol 2006;80:3675ŌĆō3678.




8. Pauly MD, Procario MC, Lauring AS. A novel twelve class fluctuation test reveals higher than expected mutation rates for influenza A viruses. Elife 2017;6:e26437.




10. Doyle JD, Chung JR, Kim SS, Gaglani M, Raiyani C, Zimmerman RK, et al. Interim estimates of 2018-19 seasonal influenza vaccine effectiveness - United States, February 2019. MMWR Morb Mortal Wkly Rep 2019;68:135ŌĆō139.



11. Rose A, Kissling E, Emborg HD, Larrauri A, McMenamin J, Pozo F, et al. Interim 2019/20 influenza vaccine effectiveness: six European studies, September 2019 to January 2020. Euro Surveill 2020;25:2000153.



12. Chittaganpitch M, Supawat K, Olsen SJ, Waicharoen S, Patthamadilok S, Yingyong T, et al. Influenza viruses in Thailand: 7 years of sentinel surveillance data, 2004-2010. Influenza Other Respir Viruses 2012;6:276ŌĆō283.


13. Simmerman JM, Chittaganpitch M, Levy J, Chantra S, Maloney S, Uyeki T, et al. Incidence, seasonality and mortality associated with influenza pneumonia in Thailand: 2005-2008. PLoS One 2009;4:e7776.



14. Tewawong N, Vichiwattana P, Korkong S, Klinfueng S, Suntronwong N, Thongmee T, et al. Evolution of the neuraminidase gene of seasonal influenza A and B viruses in Thailand between 2010 and 2015. PLoS One 2017;12:e0175655.



15. Tewawong N, Suntronwong N, Korkong S, Theamboonlers A, Vongpunsawad S, Poovorawan Y. Evidence for influenza B virus lineage shifts and reassortants circulating in Thailand in 2014-2016. Infect Genet Evol 2017;47:35ŌĆō40.


16. Tewawong N, Suntronwong N, Vichiwattana P, Vongpunsawad S, Theamboonlers A, Poovorawan Y. Genetic and antigenic characterization of hemagglutinin of influenza A/H3N2 virus from the 2015 season in Thailand. Virus Genes 2016;52:711ŌĆō715.



17. Suntronwong N, Klinfueng S, Vichiwattana P, Korkong S, Thongmee T, Vongpunsawad S, et al. Genetic and antigenic divergence in the influenza A(H3N2) virus circulating between 2016 and 2017 in Thailand. PLoS One 2017;12:e0189511.



18. Imai K, Tamura K, Tanigaki T, Takizawa M, Nakayama E, Taniguchi T, et al. Whole genome sequencing of influenza A and B viruses with the MinION sequencer in the clinical setting: a pilot study. Front Microbiol 2018;9:2748.



19. Alnaji FG, Holmes JR, Rendon G, Vera JC, Fields CJ, Martin BE, et al. Sequencing framework for the sensitive detection and precise mapping of defective interfering particle-associated deletions across influenza A and B viruses. J Virol 2019;93:e00354ŌĆōe00319.




20. Suwannakarn K, Payungporn S, Chieochansin T, Samransamruajkit R, Amonsin A, Songserm T, et al. Typing (A/B) and subtyping (H1/H3/H5) of influenza A viruses by multiplex real-time RT-PCR assays. J Virol Methods 2008;152:25ŌĆō31.


21. Tewawong N, Chansaenroj J, Klinfueng S, Vichiwattana P, Korkong S, Thongmee T, et al. Lineage-specific detection of influenza B virus using real-time polymerase chain reaction with melting curve analysis. Arch Virol 2016;161:1425ŌĆō1435.



22. World Health Organization. WHO manual on animal influenza diagnosis and surveillance. Geneva: World Health Organization, 2002. Accessed 2021 Dec 10. Available from: https://apps.who.int/iris/handle/10665/68026.

23. Ilyushina NA, Ikizler MR, Kawaoka Y, Rudenko LG, Treanor JJ, Subbarao K, et al. Comparative study of influenza virus replication in MDCK cells and in primary cells derived from adenoids and airway epithelium. J Virol 2012;86:11725ŌĆō11734.




24. Xue J, Chambers BS, Hensley SE, Lopez CB. Propagation and characterization of influenza virus stocks that lack high levels of defective viral genomes and hemagglutinin mutations. Front Microbiol 2016;7:326.



25. Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, et al. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol 2009;83:10309ŌĆō10313.




26. Korsun NS, Angelova SG, Trifonova IT, Georgieva IL, Tzotcheva IS, Mileva SD, et al. Predominance of influenza B/Yamagata lineage viruses in Bulgaria during the 2017/2018 season. Epidemiol Infect 2019;147:e76.



27. Hall T. BioEdit: an important software for molecular biology. GERF Bull Biosci 2011;2:60ŌĆō61.
28. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 2018;35:1547ŌĆō1549.



29. Kofler R, Orozco-terWengel P, De Maio N, Pandey RV, Nolte V, Futschik A, et al. PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One 2011;6:e15925.



30. Thongpan I, Suntronwong N, Vichaiwattana P, Wanlapakorn N, Vongpunsawad S, Poovorawan Y. Respiratory syncytial virus, human metapneumovirus, and influenza virus infection in Bangkok, 2016-2017. PeerJ 2019;7:e6748.




31. Hoper D, Hoffmann B, Beer M. A comprehensive deep sequencing strategy for full-length genomes of influenza A. PLoS One 2011;6:e19075.



32. Zhao J, Liu J, Vemula SV, Lin C, Tan J, Ragupathy V, et al. Sensitive detection and simultaneous discrimination of influenza A and B viruses in nasopharyngeal swabs in a single assay using next-generation sequencing-based diagnostics. PLoS One 2016;11:e0163175.



33. Kampmann ML, Fordyce SL, Avila-Arcos MC, Rasmussen M, Willerslev E, Nielsen LP, et al. A simple method for the parallel deep sequencing of full influenza A genomes. J Virol Methods 2011;178:243ŌĆō248.


34. Zhao XN, Zhang HJ, Li D, Zhou JN, Chen YY, Sun YH, et al. Whole-genome sequencing reveals origin and evolution of influenza A(H1N1)pdm09 viruses in Lincang, China, from 2014 to 2018. PLoS One 2020;15:e0234869.



35. Meinel DM, Heinzinger S, Eberle U, Ackermann N, Schonberger K, Sing A. Whole genome sequencing identifies influenza A H3N2 transmission and offers superior resolution to classical typing methods. Infection 2018;46:69ŌĆō76.



36. Xue KS, Moncla LH, Bedford T, Bloom JD. Within-host evolution of human influenza virus. Trends Microbiol 2018;26:781ŌĆō793.



37. Van Poelvoorde LAE, Saelens X, Thomas I, Roosens NH. Next-generation sequencing: an eye-opener for the surveillance of antiviral resistance in influenza. Trends Biotechnol 2020;38:360ŌĆō367.


38. Deng YM, Spirason N, Iannello P, Jelley L, Lau H, Barr IG. A simplified Sanger sequencing method for full genome sequencing of multiple subtypes of human influenza A viruses. J Clin Virol 2015;68:43ŌĆō48.


39. Cobbin JCA, Alfelali M, Barasheed O, Taylor J, Dwyer DE, Kok J, et al. Multiple sources of genetic diversity of influenza A viruses during the Hajj. J Virol 2017;91:e00096ŌĆō00017.




40. Sobel Leonard A, McClain MT, Smith GJ, Wentworth DE, Halpin RA, Lin X, et al. Deep sequencing of influenza A virus from a human challenge study reveals a selective bottleneck and only limited intrahost genetic diversification. J Virol 2016;90:11247ŌĆō11258.




41. Rondy M, Kissling E, Emborg HD, Gherasim A, Pebody R, Trebbien R, et al. Interim 2017/18 influenza seasonal vaccine effectiveness: combined results from five European studies. Euro Surveill 2018;23:18ŌĆō00086.

42. Flannery B, Chung JR, Belongia EA, McLean HQ, Gaglani M, Murthy K, et al. Interim estimates of 2017-18 seasonal influenza vaccine effectiveness - United States, February 2018. MMWR Morb Mortal Wkly Rep 2018;67:180ŌĆō185.



43. Centers for Disease Control and Prevention. Summary of the 2017-2018 Influenza Season, 2019. Atlanta, GA: Centers for Disease Control and Prevention, 2019. Accessed 2021 Dec 10. Available from: https://www.cdc.gov/flu/about/season/flu-season-2017-2018.htm.

44. World Health Organization. Recommended composition of influenza virus vaccines for use in the 2018 southern hemisphere influenza season. Geneva: World Health Organization, 2017. Accessed 2021 Dec 10. Available from: https://www.who.int/influenza/vaccines/virus/recommendations/201709_recommendation.pdf?ua=1.
45. Suntronwong N, Klinfueng S, Korkong S, Vichaiwattana P, Thongmee T, Vongpunsawad S, et al. Characterizing genetic and antigenic divergence from vaccine strain of influenza A and B viruses circulating in Thailand, 2017-2020. Sci Rep 2021;11:735.




46. Belanov SS, Bychkov D, Benner C, Ripatti S, Ojala T, Kankainen M, et al. Genome-wide analysis of evolutionary markers of human influenza A(H1N1)pdm09 and A(H3N2) viruses may guide selection of vaccine strain candidates. Genome Biol Evol 2015;7:3472ŌĆō3483.



47. Elderfield RA, Watson SJ, Godlee A, Adamson WE, Thompson CI, Dunning J, et al. Accumulation of human-adapting mutations during circulation of A(H1N1)pdm09 influenza virus in humans in the United Kingdom. J Virol 2014;88:13269ŌĆō13283.




48. Sarmah K, Borkakoty B, Sarma K, Hazarika R, Das PK, Jakharia A, et al. Genetic variations of the Hemagglutinin gene of Pandemic Influenza A (H1N1) viruses in Assam, India during 2016. 3 Biotech 2018;8:408.




49. Ramos AP, Herrera BA, Ramirez OV, Garcia AA, Jimenez MM, Valdes CS, et al. Molecular and phylogenetic analysis of influenza A H1N1 pandemic viruses in Cuba, May 2009 to August 2010. Int J Infect Dis 2013;17:e565ŌĆōe567.


50. Roubidoux EK, Carreno JM, McMahon M, Jiang K, van Bakel H, Wilson P, et al. Mutations in the hemagglutinin stalk domain do not permit escape from a protective, stalk-based vaccine-induced immune response in the mouse model. mBio 2021;12:e03617ŌĆōe03620.




51. Jimenez-Jorge S, Pozo F, de Mateo S, Delgado-Sanz C, Casas I, Garcia-Cenoz M, et al. Influenza vaccine effectiveness in Spain 2013/14: subtype-specific early estimates using the cycEVA study. Euro Surveill 2014;19:20727.

52. European Centre of Disease Prevention and Control (ECDC). Surveillance report. Influenza virus characterization. Summary Europe. Solna: European Centre of Disease Prevention and Control, 2016. Accessed 2021 Dec 10. Available from: https://www.ecdc.europa.eu/sites/default/files/media/en/publications/Publications/influenza-virus-characterisation-may-2016.pdf.
53. Cheng AC, Subbarao K. Epidemiological data on the effectiveness of influenza vaccine: another piece of the puzzle. J Infect Dis 2018;218:176ŌĆō178.


54. Jones S, Prasad R, Nair AS, Dharmaseelan S, Usha R, Nair RR, et al. Whole-genome sequences of influenza A(H1N1)pdm09 virus isolates from Kerala, India. Genome Announc 2017;5:e00598ŌĆō00517.




55. Al Khatib HA, Al Maslamani MA, Coyle PV, Thompson IR, Farag EA, Al Thani AA, et al. Inter- versus intra-host sequence diversity of pH1N1 and associated clinical outcomes. Microorganisms 2020;8:133.



56. Clark AM, Nogales A, Martinez-Sobrido L, Topham DJ, DeDiego ML. Functional evolution of influenza virus NS1 protein in currently circulating human 2009 pandemic H1N1 viruses. J Virol 2017;91:e00721ŌĆō00717.




57. Krug RM. Functions of the influenza A virus NS1 protein in antiviral defense. Curr Opin Virol 2015;12:1ŌĆō6.



58. Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3'end formation of cellular pre-mRNAs. Mol Cell 1998;1:991ŌĆō1000.


59. Komissarov A, Fadeev A, Sergeeva M, Petrov S, Sintsova K, Egorova A, et al. Rapid spread of influenza A(H1N1)pdm09 viruses with a new set of specific mutations in the internal genes in the beginning of 2015/2016 epidemic season in Moscow and Saint Petersburg (Russian Federation). Influenza Other Respir Viruses 2016;10:247ŌĆō253.




60. Mao H, Sun Y, Chen Y, Lou X, Yu Z, Wang X, et al. Co-circulation of influenza A(H1N1), A(H3N2), B(Yamagata) and B(Victoria) during the 2017-2018 influenza season in Zhejiang Province, China. Epidemiol Infect 2020;148:e296.



61. Boonnak K, Mansanguan C, Schuerch D, Boonyuen U, Lerdsamran H, Jiamsomboon K, et al. Molecular characterization of seasonal influenza A and B from hospitalized patients in Thailand in 2018-2019. Viruses 2021;13:977.



62. Jagadesh A, Krishnan A, Nair S, Sivadas S, Arunkumar G. Genetic characterization of hemagglutinin (HA) gene of influenza A viruses circulating in Southwest India during 2017 season. Virus Genes 2019;55:458ŌĆō464.



63. Wan H, Gao J, Yang H, Yang S, Harvey R, Chen YQ, et al. The neuraminidase of A(H3N2) influenza viruses circulating since 2016 is antigenically distinct from the A/Hong Kong/4801/2014 vaccine strain. Nat Microbiol 2019;4:2216ŌĆō2225.




- TOOLS
-
METRICS

-
- 3 Crossref
- 0 Scopus
- 5,129 View
- 244 Download







