1. Osterberg L, Blaschke T. Adherence to medication. N Engl J Med 2005;353:487ŌĆō497. PMID:
16079372.


2. Zwikker HE, van den Bemt BJ, Vriezekolk JE, van den Ende CH, van Dulmen S. Psychosocial predictors of non-adherence to chronic medication: systematic review of longitudinal studies. Patient Prefer Adherence 2014;8:519ŌĆō563. PMID:
24851043.



3. Jin J, Sklar GE, Min Sen Oh V, Chuen Li S. Factors affecting therapeutic compliance: A review from the patient's perspective. Ther Clin Risk Manag 2008;4:269ŌĆō286. PMID:
18728716.



5. McCrae RR, Costa PT Jr. Personality in Adulthood: A Five-Factor Theory Perspective. New York: Guilford Press, 2003.
6. Jerant A, Chapman B, Duberstein P, Robbins J, Franks P. Personality and medication non-adherence among older adults enrolled in a six-year trial. Br J Health Psychol 2011;16(Pt 1):151ŌĆō169. PMID:
21226789.



7. Axelsson M, Brink E, Lundgren J, L├Čtvall J. The influence of personality traits on reported adherence to medication in individuals with chronic disease: an epidemiological study in West Sweden. PLoS One 2011;6:e18241. PMID:
21464898.



8. Kim HN, Roh SJ, Sung YA, Chung HW, Lee JY, Cho J,
et al. Genome-wide association study of the five-factor model of personality in young Korean women. J Hum Genet 2013;58:667ŌĆō674. PMID:
23903073.


9. Terracciano A, Sanna S, Uda M, Deiana B, Usala G, Busonero F,
et al. Genome-wide association scan for five major dimensions of personality. Mol Psychiatry 2010;15:647ŌĆō656. PMID:
18957941.


10. Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ,
et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet 2009;41:527ŌĆō534. PMID:
19396169.


11. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D,
et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559ŌĆō575. PMID:
17701901.



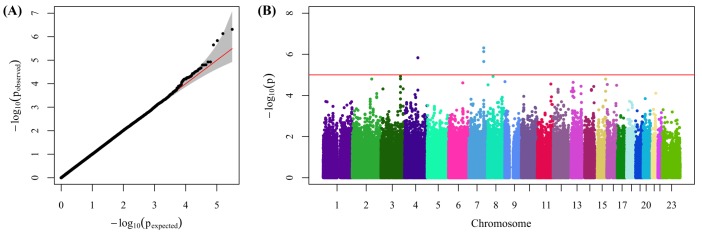
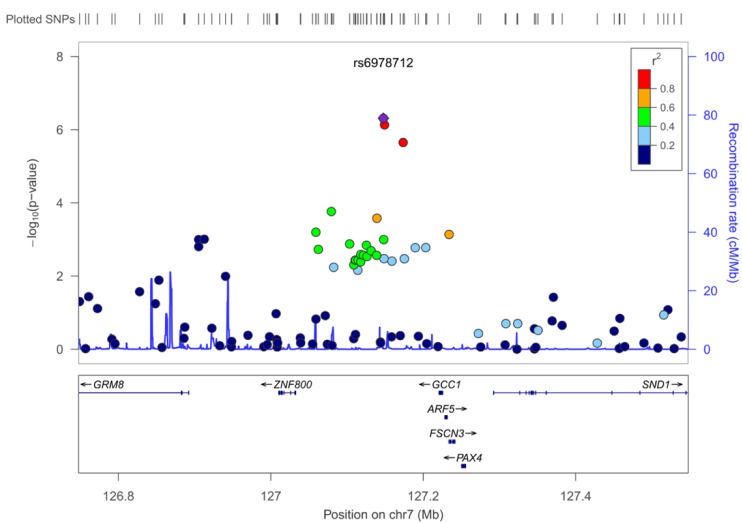
13. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP,
et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010;26:2336ŌĆō2337. PMID:
20634204.



14. Briesacher BA, Andrade SE, Fouayzi H, Chan KA. Comparison of drug adherence rates among patients with seven different medical conditions. Pharmacotherapy 2008;28:437ŌĆō443. PMID:
18363527.



15. Lehrmann E, Colantuoni C, Deep-Soboslay A, Becker KG, Lowe R, Huestis MA,
et al. Transcriptional changes common to human cocaine, cannabis and phencyclidine abuse. PLoS One 2006;1:e114. PMID:
17205118.



16. Malhotra AK, Zhang JP, Lencz T. Pharmacogenetics in psychiatry: translating research into clinical practice. Mol Psychiatry 2012;17:760ŌĆō769. PMID:
22083729.



17. Bush WS, Moore JH. Chapter 11: Genome-wide association studies. PLoS Comput Biol 2012;8:e1002822. PMID:
23300413.



18. Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA 2008;299:1335ŌĆō1344. PMID:
18349094.


19. Horne R, Weinman J. Self-regulation and self-management in asthma: exploring the role of illness perceptions and treatment beliefs in explaining non-adherence to preventer medication. Psychol Health 2002;17:17ŌĆō32.

20. Morisky DE, Ang A, Krousel-Wood M, Ward HJ. Predictive validity of a medication adherence measure in an outpatient setting. J Clin Hypertens (Greenwich) 2008;10:348ŌĆō354. PMID:
18453793.



21. Kjer-Nielsen L, Teasdale RD, van Vliet C, Gleeson PA. A novel Golgi-localisation domain shared by a class of coiled-coil peripheral membrane proteins. Curr Biol 1999;9:385ŌĆō388. PMID:
10209125.