Genome-Wide Association Study of Orthostatic Hypotension and Supine-Standing Blood Pressure Changes in Two Korean Populations
Article information
Abstract
Orthostatic hypotension (OH) is defined by a 20-mm Hg difference of systolic blood pressure (dtSBP) and/or a 10-mm Hg difference of diastolic blood pressure (dtDBP) between supine and standing, and OH is associated with a failure of the cardiovascular reflex to maintain blood pressure on standing from a supine position. To understand the underlying genetic factors for OH traits (OH, dtSBP, and dtDBP), genome-wide association studies (GWASs) using 333,651 single nucleotide polymorphisms (SNPs) were conducted separately for two population-based cohorts, Ansung (n = 3,173) and Ansan (n = 3,255). We identified 8 SNPs (5 SNPs for dtSBP and 3 SNPs for dtDBP) that were repeatedly associated in both the Ansung and Ansan cohorts and had p-values of <1 × 10-5 in the meta-analysis. Unfortunately, the SNPs of the OH case control GWAS did not pass our p-value criteria. Four of 8 SNPs were located in the intergenic region of chromosome 2, and the nearest gene (CTNNA2) was located at 1 Mb of distance. CTNNA2 is a linker between cadherin adhesion receptors and the actin cytoskeleton and is essential for stabilizing dendritic spines in rodent hippocampal neurons. Although there is no report about the function in blood pressure regulation, hippocampal neurons interact primarily with the autonomic nervous system and might be related to OH. The remaining SNPs, rs7098785 of dtSBP trait and rs6892553, rs16887217, and rs4959677 of dtDBP trait were located in the PIK3AP1 intron, ACTBL2-3' flanking, STAR intron, and intergenic region, respectively, but there was no clear functional link to blood pressure regulation.
Introduction
Orthostasis causes a gravitational shift in circulating blood from the intra-thoracic space to lower extremities and reduces venous return to the heart. This transient increase in sympathetic tone and decrease in parasympathetic tone and these normal cardiovascular reflexes cause an increase in heart rate and vascular resistance, restoring normal blood pressure (BP) and cardiac output [1]. Orthostatic hypotension (OH) is the failure of the cardiovascular reflexes to maintain BP on standing from a supine position [2]. The Malmö Preventive Project reported that individuals with OH had significantly increased all-cause mortality (hazard ratio, 1.4 to 1.6) [3].
OH has been defined by the international consensus as a decrease in systolic BP ≥ 20 mm Hg and/or a decrease in diastolic BP ≥ 10 mm Hg within 3 mintues of standing [4]. The prevalence of OH varies from 5% to 30%, depending greatly on the population being studied [5-7]. Aging is associated with a definite increase in the prevalence of OH and was reviewed in Benvenuto and Krakoff [2]. In addition to the aging effects, OH is more prevalent in hypertensive [8, 9], neurological [10], and diabetic patients [11, 12]. The all-cause mortality rate was higher in patients with OH than in those without OH [6, 7]
We have been conducting an epidemiology study of OH as part of the Korean Genome and Epidemiology Study (KoGES) and previously reported that middle-aged adults (40-70 years), enrolled from 2001 to 2002, had a 12.3% overall prevalence of OH and that the OH frequency increased significantly with age, from 6.4% in those aged 40-44 years to 23.1% in those aged 65-69 (p < 0.001) [9]. In addition, the study confirmed the correlations between OH and hypertension, body mass index (BMI), and type 2 diabetes [9].
The genetic contribution to OH can be hypothesized that the frequency of OH in families with a history of essential hypertension is higher than in the general population [4]. A genetic association study was conducted by Luo et al. [13], and they identified a functional variant of NEDD4L (epithelial sodium channel E3-ubiquitin ligase).
To identify genetic factors associated with OH trait in Koreans, we conducted genome-wide association studies (GWASs) for OH case controls using genome-wide single nucleotide polymorphism (SNP) genotypes and also tested supine and standing BPs as quantitative traits.
Methods
Study subjects
The study subjects consisted of two population-based cohorts, Ansung and Ansan, which have been conducted as part of the KoGES. The phenotype of the cohort population was previously described [14]. Briefly, the subjects came from the Ansung and Ansan cities, located in Gyeonggi-do province near Seoul, Korea. Written informed consent was obtained from all participants, and this research project was approved by the institutional review board in the Korea National Institute of Health (KNIH).
Subjects with genotype accuracies below 98% and high missing genotype call rates (≥4%), high heterozygosity (>30%), or inconsistency in sex were excluded from subsequent analyses. Individuals who had a tumor were excluded, as were related individuals whose estimated identity-by-state values were high (>0.80). After these quality control steps, 8,842 samples were selected.
Clinical characteristics
BP was measured 3 times in the supine position. Before the first measurement, participants rested for 5 minutes, and the 3 measurements were taken in one arm showing the higher BP at least 3 minutes apart. After the supine position measurement, the BP was measured soon after standing. Two of 8,842 subjects did not undergo BP measurement in the standing posture. Also, 1,291 individuals who had been treated with anti-hypertensive drug therapy and 1,114 self-reported diabetic patients or diagnosed diabetic patients by KoGES were excluded. Ultimately, 3,255 individuals from Ansan and 3,173 individuals from Ansung were used for the GWASs. Other cardiovascular risk factors, such as cholesterol level and fasting glucose level, were measured from blood samples after overnight fasting.
Study genotypes
The genotyping of the cohort population was previously described for the Korea Associated Resource (KARE) study [14]. Most DNA samples were isolated from the peripheral blood of participants and genotyped using the Affymetrix Genomewide Human SNP array 5.0 (Affymetrix, Inc., Santa Clara, CA, USA). The quality control steps of genotypes have been described elsewhere [14]. Briefly, the accuracy of the genotyping was determined by Bayesian Robust Linear Modeling using the Mahalanobis Distance (BRLMM) genotyping algorithm [15]. Consequently, 333,651 SNPs had a missing genotype call rate below 0.1, a minor allele frequency (MAF) greater than 0.01, and no deviation from Hardy-Weinberg equilibrium (HWE) (p > 1 × 10-6). To examine the population stratification, multidimensional scaling analysis and principal component analysis were performed using 44,724 pruned SNP markers [14].
Statistical analysis
The orthostatic BP changes were calculated by deltaBP = mean supine position BP - standing position BP. Delta systolic blood pressures (dtSBP), delta diastolic blood pressure (dtDBP), and OH cases (dtSBP ≥ 20 and/or dtDBP ≥ 10 [16]) were analyzed by linear or logistic regression, controlling for covariates, such as age, sex, BMI, and systolic BP. Statistical analyses were performed using PLINK version 1.07, using default options [17] and SPSS version 15.0 (SPSS Inc., Chicago, IL, USA). The GWAS were conducted for Ansan and Ansung separately. The Ansan GWAS and Ansung replication study results were combined by inverse-variance meta-analysis method, assuming fixed effects, with Cochran's Q test used to assess between-study heterogeneity [18]. The selected SNPs had a common MAF (>0.05) and the population heterogeneity (p > 0.05). The asymptotic HWE tests were conducted using PLINK, and all the reported p-values were two-sided.
Results
Clinical characteristics
Table 1 describes the clinical characteristics of OH case and control subjects in the Ansan and Ansung cohorts; 311 participants (7.6%) and 496 participants (14.3%) were found to have OH in Ansan and Ansung, respectively. The gender distributions of OH differed significantly in both cohorts, and more male OH in Ansung and less male OH in Ansan were observed. The age of OH in Ansung (58.8 ± 7.8 years) was older than that in Ansan (51.5 ± 8.3 years), reflecting the higher prevalence of OH in Ansung.

Clinical characteristics of orthostatic hypotension case controls in Ansan and Ansung cohorts phenotype
The BMI was significantly smaller in Ansung OH cases than controls, but it was not significant in Ansan. While the mean supine BP was significantly higher in OH than controls, the standing BP was significantly lower in OH than controls.
Genome-wide association studies
GWAS were conducted for each cohort, and the target phenotypes were dtSBP, dtDBP, and OH. The GWAS results of each cohort were combined by meta-analyses, and quantile-quantile plots and Manhattan plots are depicted in Fig. 1.

Quantile-quantile plots and Manhattan plots of genomewide association studies for delta systolic blood pressure (A, D), delta diastolic blood pressure (B, E), and orthostatic hypotension (C, F).
All detected SNPs are described in Table 2. Five SNPs for dtSBP trait and three SNPs for dtDBP trait passed our criteria (replicated in both cohorts [Ansung and Ansan] and p < 1 × 10-5), but unfortunately, the SNPs of the OH case control GWAS did not pass our p-value criteria. Among the significant SNPs, four SNPs (rs6736587, rs6756959, rs6715049, and rs17020502) were in the high linkage disequilibrium status (r2 > 0.9) and were including the most significant SNP, rs6736587 (meta-analysis p-value, 5.3 × 10-8). The region of four SNPs was located in the intergenic region of chromosome 2, and the nearest gene (CTNNA2) was located at 1 Mb of distance. The remaining SNPs revealed a suggestive association (p < 1 × 10-5) with dtSBP at rs7098785 (PIK3AP1 intron) and with dtDBP at rs6892553, rs16887217, and rs4959677, which were located in the ACTBL2 3' flanking, steroidogenic acute regulatory protein (STAR) intron, and intergenic regions (chromosome 6p 25.2), respectively.
Discussion
These normal baro-reflex responses cause an increase in heart rate and vascular resistance, restoring normal BP and cardiac output [1]. OH is the failure of the cardiovascular reflexes to maintain BP on standing from a supine position [2]. Also, the frequency of OH in families with a history of essential hypertension may be higher than in the general population [4]. Therefore, the identification of genetic factor for orthostasis will help us understand OH mechanisms.
The signal plot for the most significant SNP (rs6736587) is depicted in Fig. 2; the region was intergenic and a functional gene was located 1 Mbp upstream. Interestingly, this region is known as the chromosome 2p12-11.2 deletion syndrome region, and several cases report mental or psychomotor retardation [19, 20]. There was only one gene, CTNNA2 (or known as alpha-N-catenin), around ±1 Mbp. CTNNA2 is a linker between cadherin adhesion receptors and the actin cytoskeleton and is essential for stabilizing dendritic spines in rodent hippocampal neurons [21]. Although there is no report about function in BP regulation, hippocampal neurons interact primarily with the autonomic nervous system. Therefore, the CTNNA2 gene may function in orthostatic BP regulation.
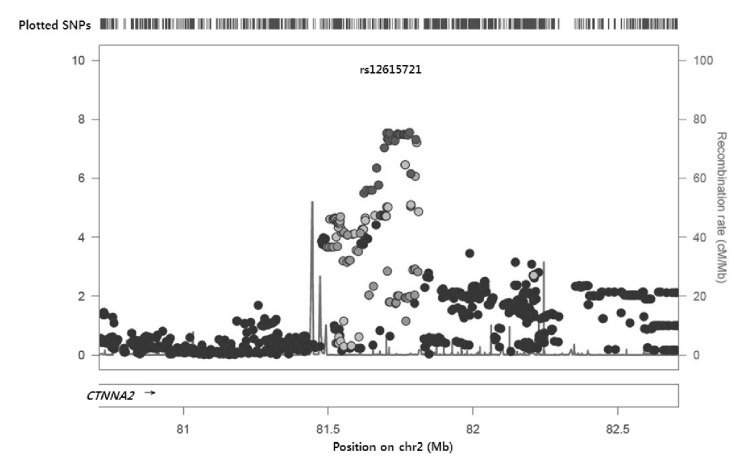
Signal plot of the most significant association with delta systolic blood pressure. SNP, single nucleotide polymorphism.
Among the other SNPs, rs7098785 was located in the phosphoinositide-3-kinase adaptor protein 1 (PIK3AP1) gene. PIK3AP1 is involved in B-cell development [22]. Interestingly, in silico analyses by the UCSC genome browser (http://genome.ucsc.edu/) indicated that the SNP was located in a DNase I hypersensitive region and in the binding site of transcription factors (MEF2C, BATF, MEF2A). Therefore, the rs7098785 SNP might have a regulatory function of PIK3AP1 expression.
The remaining three SNPs of dtDBP were located in the ACTBL2 (beta-actin like protein 2) 3' flanking region, STAR intron, and intergenic region (chromosome 6p 25.2). The ACTBL2 protein is involved in various types of cell motility [23], and STAR is involved in the regulation of steroid hormone synthesis [24].
Our study was the first trial to identify genetic variants for orthostatic BP traits in Koreans. We hope that our finding could help us understand the underlying mechanism of orthostatic BP changes and related diseases.