Metabolic Pathways Associated with Kimchi, a Traditional Korean Food, Based on In Silico Modeling of Published Data
Article information
Abstract
Kimchi is a traditional Korean food prepared by fermenting vegetables, such as Chinese cabbage and radishes, which are seasoned with various ingredients, including red pepper powder, garlic, ginger, green onion, fermented seafood (Jeotgal), and salt. The various unique microorganisms and bioactive components in kimchi show antioxidant activity and have been associated with an enhanced immune response, as well as anti-cancer and anti-diabetic effects. Red pepper inhibits decay due to microorganisms and prevents food from spoiling. The vast amount of biological information generated by academic and industrial research groups is reflected in a rapidly growing body of scientific literature and expanding data resources. However, the genome, biological pathway, and related disease data are insufficient to explain the health benefits of kimchi because of the varied and heterogeneous data types. Therefore, we have constructed an appropriate semantic data model based on an integrated food knowledge database and analyzed the functional and biological processes associated with kimchi in silico. This complex semantic network of several entities and connections was generalized to answer complex questions, and we demonstrated how specific disease pathways are related to kimchi consumption.
Introduction
Kimchi is a salty, fermented preparation of cabbage that is widely consumed in the traditional Korean diet. Certain types of kimchi are prepared by mixing salted cabbage with spicy ingredients, such as red pepper powder, garlic, and ginger, while others are prepared without red chili pepper or are soaked in a savory liquid. Kimchi is prepared by mixing cabbage well with other vegetables, fish seasoning, and salt, and packing it into jars. The various unique microorganisms [1] and bioactive components present in kimchi have shown antioxidant activity and have been associated with an enhanced immune response, as well as anti-cancer and anti-diabetic effects. For example, components produced during the fermentation process show anti-cancer effects via the inhibition of the Ras oncogene signaling pathway [2], and anti-atherosclerotic effects are seen via a reduction in low-density lipoprotein-cholesterol oxidation [3456]. During the fermentation process, bacteria in the environment, predominantly from the genera Lactobacilli, Leuconostoc, and Weissella, produce byproducts that contribute to the pungent flavor of kimchi and include beneficial bioactive compounds [78].
In particular, the red chili pepper, the component of kimchi that makes it spicy, is a potent antioxidant due to its high vitamin C levels and capsaicin content. Furthermore, red pepper inhibits the decay of microorganisms and prevents food from spoiling. The physiological effects of kimchi thus originate from bioactive compounds present in its ingredients, and the functions of these are amplified by the fermentation process. Therefore, the health-promoting, probiotic, and functional properties of kimchi can be optimized by manipulating the types and amounts of ingredients, and by using appropriate probiotic starters and kimchi preparation methods such as fermentation. Optimally produced kimchi can be a very healthy food [9].
However, the available genome, biological pathway, and related disease data are still insufficient to explain the health benefits of kimchi. An integrated database of these data is not available, because it is difficult to control for the heterogeneous data types. Furthermore, biological processes are interrelated on many levels and their regulation presents complexity. These interactions must be understood in detail to successfully manage risks in the development of biological products. Evidence-based information on kimchi should be provided by both public and proprietary information sources, including public databases, experimental results from individual studies, text-mining analyses, and knowledge management systems. The model integrating this data would represent the accumulated knowledge in this domain that could be browsed and mined interactively. In this study, we have constructed the proper semantic data model and extracted integrated information on kimchi's nutritional content, and its effects on physiology and disease, based on analyses of the functional and biological characteristics of kimchi in the scientific literature.
Methods
Semantic modeling
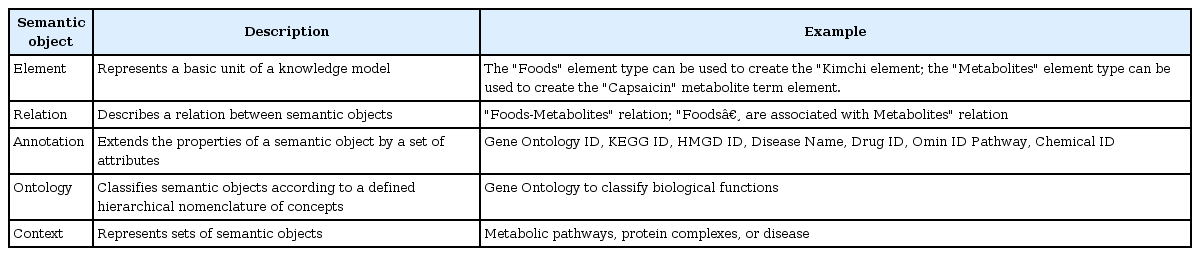
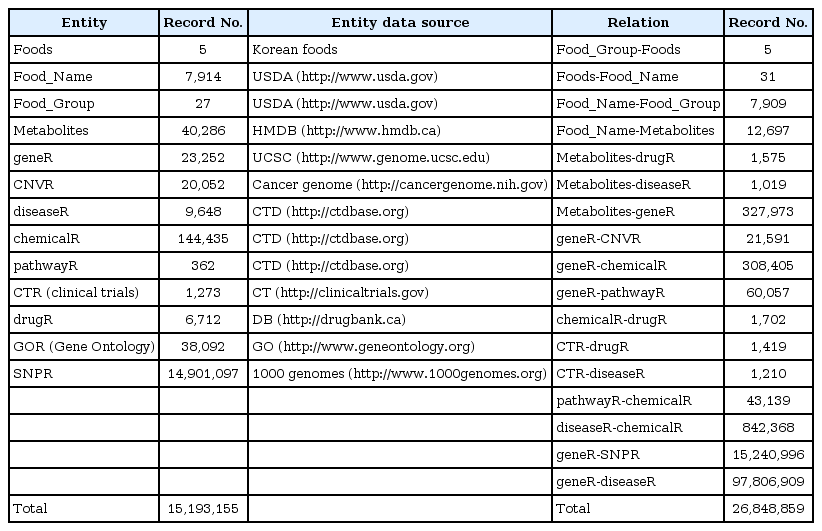
We used a semantic tool for knowledge base management, BioXM (Biomax Information AG, Munchen, Germany), which was developed for object-oriented semantic integration. In this approach, semantically identical objects and the associations between them are identified and mapped based on data and descriptive meta-information [10]. This primarily involves the mapping of biological objects and descriptive data from literature to common objects like genes, diseases, or chemical. Associations between entities are mapped as relations (e.g., chemical X is associated with disease A) and object relation information is contextually structured (e.g., gene B is regulated negatively/positively by chemical X). Using objects as nodes and relations as edges, a “semantic network” can be generated that provides information on the connections between participating objects. Various elementary concepts, such as elements, relations, and annotations, are defined in Fig. 1 and the study workflow is illustrated in Fig. 2. We modified this complex semantic network to detect connections, extract patterns, and answer complex questions, as is shown in Fig. 3. To access the data easily, we used the BioXM tool, which provides visual browsing of the knowledge network (Fig. 3), as well as data mapping and query wizards (Fig. 4) that automatically adapt to the changes of a given data model. This system enabled us to create knowledge networks with flexible workflows for handling data and information types.
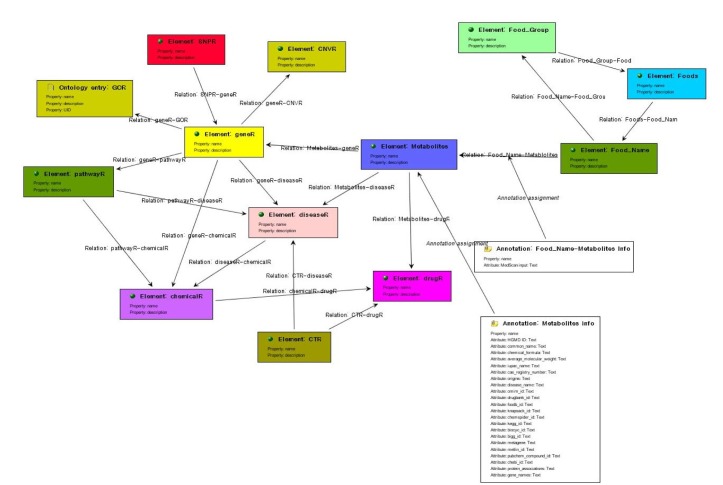
Korean Food Standard Reference semantic modeling. Semantic data modeling for integration of the biological relationships among various semantic elements, such as foods, nutrients, genes, chemicals, diseases, and pathways, derived from public databases.

The study workflow. SNP, single nucleotide polymorphism; KEGG, Kyoto Encyclopedia of Genes and Genomes.
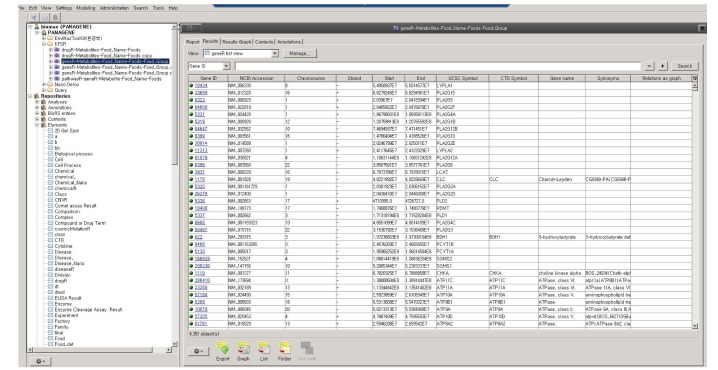
Kimchi-related genes determined by semantic modeling of data in the Korean Food Standard Reference (KFSR). A total of 4,351 genes were extracted from the KFSR. The blue box indicates a query on how genes are extracted by semantic modeling.
Data collection by text-mining and data analysis
Pathway Studio software 9.0 (Elsevier, Atlanta, GA, USA) was used to collect information on biological processes and diseases related to kimchi, based on information in the Korean Food Standard Reference (KFSR). Pathway Studio software allows the automatic extraction of regulatory and physical interactions from MEDLINE abstracts using a natural-language-processing technology called MedScan [11]. Interactions extracted by MedScan, which contain a formalized set of relationships, are imported into the Pathway Studio database and analyzed further using data-mining tools for knowledge inference [12]. Since MedScan keeps the reference of the original article with information on the extracted interaction, it also allows a quick review of extracted facts and identification of relevant publications. Thus, it facilitated our pathway reconstruction by selecting the appropriate interactions. All interactions found were manually curated, and only validated interactions were included in the pathways.
Results
Semantic data modeling
Elements and relations were maintained and linked by relation and object. This semantic model enabled elements to be mapped to known elements. Relations between biological data are used to efficiently extract interactions of interest. The semantic modeling in this study was configured in the generic BioXM knowledge management environment to create a knowledge base for this translational systems biology approach. We constructed semantic networks for the discovery of a network biomarker using BioXM software (Biomax Information AG), which is a customizable knowledge base management tool for scientific data. The graphical data model is shown in Fig. 1. As shown in Table 1, we created elements such as “Foods,” “Foods Group,” “Food Name,” “Metabolites,” and nine other elements to describe the available biological knowledge. Information on the interconnection of elements is captured in relations such as “is related to” for “Foods” or “regulates” for a metabolite that regulates expression of a gene. This network consisted of 13 elements and 20 relations. The semantic network provided information on connections between included objects instantly. Therefore, this integrated semantic data model was used to create new knowledge networks with flexible workflows.
Data configuration
This semantic model enabled us to integrate existing public databases and experimental data derived from the literature. To populate the knowledge base with data from public databases, we manually generated mappings in import-templates. A graphical wizard for import-template generation provided a selection of possible import options and mappings.
Construction of the KFSR
The United States Department of Agriculture (USDA) food and nutrient databases provide the basic infrastructure for food and nutrition research, nutrition monitoring, policy, and dietary practice. The databases date back to 1892 and they are unique, as the only databases available in the public domain that perform these functions. The Nutrient Data Laboratory develops and maintains the USDA Standard Reference and related data products [13]. The Standard Reference is the major source of food composition data in the United States. It provides the foundation for most food composition databases used in food policy, research, dietary practice, and nutrition monitoring. The USDA databases were used to construct the KFSR. This product is the basis of numerous other applications that address nutrients in foods. The database contains information on agricultural commodities, as well as data on formulated and processed foods. Currently, the database contains values for up to 140 different food components, including nitrogen (protein), total fat, carbohydrates, and moisture, as well as individual fatty acids, amino acids, vitamins, and minerals in >7,500 foods. We have added five traditional Korean foods, i.e., kimchi, bulgogi, bibimbap, doenjang, and gochujang. To construct a useful KFSR, we integrated 11 public databases as shown in Table 2, with a total of 26,848,859 records.
Data analysis of kimchi from the knowledge base
We found a total 4,351 genes strongly associated with kimchi's secondary metabolites using the KFSR, as shown in Fig. 3. First, 1,237 kimchi metabolites were extracted form >7,500 foods in KFSR and 4,351 genes were associated with 1,237 kimchi metabolites. Through the semantically integrated datasets, we discovered genes that were associated with kimchi metabolites. This network is shown in Fig. 4. We used the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway to classify genes by metabolic function. The genes in Table 3 are classified into six categories, and 309 of these genes were found to be involved in lipid metabolism. At first, we initiated a cardiovascular disease network analysis related to kimchi using 30 genes. Of these, 27 were directly connected by interactions such as regulation, promoter binding, direct regulation, protein modification, and miRNA regulation. Genes like ADCY5, ADCY1, ADCY2, GNAS, CTNNB1, EDN1, and RAC1 were the most highly connected nodes with node degrees of ≥10 (p < 0.001) (Fig. 5A). Adenyl cyclases (ADCYs) are critical regulators of metabolic and cardiovascular function, and ADCY5 is increased in hearts with pressure-overloaded left ventricular hypertrophy [1415]. In addition, guanine nucleotide-binding protein Gs (GNAS) overexpression results in increased sensitivity to apoptotic stimulation and cardiomyopathy [16], as shown in Fig. 4A. In addition, GNAS leads to the activation of β-catenin which is a multifunctional protein coded by the CNNB1 gene. Although there has long been speculation on the association between kimchi and heart disease, Lee et al. [16] demonstrated that kimchi may not adversely affect blood pressure and cardiac function.

Functional classifications of genes extracted by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway map IDs
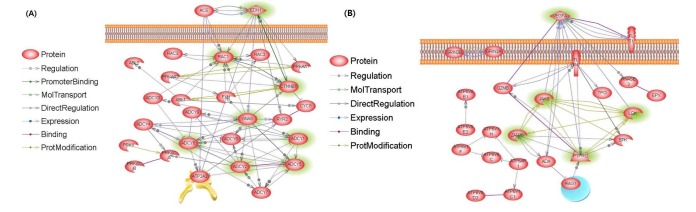
The relationship of kimchi to a disease network. (A) Cardiovascular disease. (B) Immune disease. The genes associated with kimchi nutrients are highlighted in green.
Kimchi is effective in enhancing immune function [31819]; however, the scientific evidence to support this claim has been insufficient. In this work, we performed an immune disease network analysis related to kimchi, based on public information and using 39 selected genes. Twenty-three genes were directly connected to each other by interactions such as regulation, promoter binding, direct regulation, protein modification, and miRNA regulation. In particular, PTPRC, LCK, JAK3, ZAP70, and VEGFA were the most highly connected nodes with node degrees of ≥7 (p < 0.001) (Fig. 5B). Specifically, PTPRC and JAK3 were found to regulate innate immune responses [202122]. This analytic information database, the KFSR, may suggest ways to analyze and more effectively use existing scientific data.
Discussion
For a long time, many nutritionists have believed, based on metabolomics data, that well-fermented kimchi has numerous anti-biotic activities [2324]. Recently, this has led to a significant increase in kimchi export. Kimchi has gradually gained popularity, even among foreigners, and it has become a globally recognized food. However, the genome, biological pathway, and related disease data are still insufficient to explain the health benefits of kimchi. Therefore, we investigated, in silico, a kimchi network based on public information using network-based systems biology tools, including BioXM and Pathway Studio. The in silico network data used in our study were based on published literature. With a long-term aim to search for the metabolic pathways affected by kimchi and kimchi's effects on various diseases, we first re-analyzed the data for better comparability. Then, a number of specific biomolecular networks for kimchi were built in various ways. Genes analyzed in Pathway Studio were shown to be key players in various cellular pathways and mechanisms, such as inflammation, cell proliferation, and immunity, which could be altered by underlying causes, such as disease Moreover, these biological processes were also related to obesity. This interpretation supports the view that there are common, underlying markers for targeted biological processes. Our results should be considered only the initial effort in understanding the interrelated characteristics of the highly complex metabolic pathway of kimchi. Experimental studies will also be needed to verify this in silico kimchi network.
Acknowledgments
This work was supported by a grant from the Korea Food Research Institute.