 |
 |
- Search
| Genomics Inform > Volume 21(1); 2023 > Article |
|
Abstract
The fishes of the Chiasmodontidae family, known as swallower fishes, are species adapted to live in deep seas. Several studies have shown the proximity of this family to Tetragonuridae and Amarsipidae. However, the phylogenetic position of this clade related to other Pelagiaria groups remains uncertain even when phylogenomic studies are employed. Since the low number of published mitogenomes, our study aimed to assemble six new mitochondrial genomes of Chiasmodontidae from database libraries to expand the discussion regarding the phylogeny of this group within Scombriformes. As expected, the composition and organization of mitogenomes were stable among the analyzed species, although we detected repetitive sequences in the D-loop of species of the genus Kali not seen in Chiasmodon, Dysalotus, and Pseudoscopelus. Our phylogeny incorporating 51 mitogenomes from several families of Scombriformes, including nine chiasmodontids, recovered interfamilial relationships well established in previous studies, including a clade containing Chiasmodontidae, Amarsipidae, and Tetragonuridae. However, phylogenetic relationships between larger clades remain unclear, with disagreements between different phylogenomic studies. We argue that such inconsistencies are not only due to biases and limitations in the data but mainly to complex biological events in the adaptive irradiation of Scombriformes after the Cretaceous-Paleogene extinction event.
The Chiasmodontidae family comprises 36 species of marine fishes belonging to four genera with a worldwide distribution [1]. This group, traditionally known as swallowers fishes or snake tooth fishes, have mesopelagic and bathypelagic habits living below 200 m depth [2]. As a result of the selective pressures of the environment with few food resources, these fish are known for their small size [2] and expandable stomachs, allowing the ingestion of prey much larger than the animal itself [3].
Interfamilial relationships of Chiasmodontidae with other groups are uncertain. Different phylogenetic hypotheses were postulated in recent years [4-8]. Miya et al. [4] included Chiasmodontidae and 14 other families in the Pelagiaria clade, highlighting the high support on the monophyly of the group. In addition, they argued that Amarsipidae and other Stromateoidei belong to the same clade.
Like Chiasmodontidae and other Pelagiaria groups, Stromateoidei was part of the order Perciformes in the past, until Nelson et al. [9] included Stromateoidei and Scombroidei in a new order called Scombriformes. Studies before Nelson et al. [9] had already shown that both Stromateoidei and Scombroidei suborders did not correspond to natural groupings [4-5,10,11]. Considering the phylogenetic relationships of most families traditionally allocated to these suborders with other groups within the Pelagiaria clade, Betancur et al. [5] included Chiasmodontidae and seven other Pelagiaria families in Scombriformes. Although Betancur et al. [5] recognized that interfamilial relationships of Scombriformes were uncertain, genomic studies have demonstrated a strong phylogenetic relationship between Chiasmodontidae with Tetragonuridae [4-8,12] and Amarsipidae [7,8].
To date, only the mitochondrial genomes of Chiasmodon harteli (unpublished data), Chiasmodon asper, Dysalotus alcocki, and Kali indica of the Chiamodontidae family have been published [4]. However, only K. indica, D. alcoki, and C. harteli have their mitogenomes deposited in the GenBank (the accession number provided for C. asper corresponds to the mitogenome of Champsodon cf. snyderi in the NCBI). This number is very small compared to the related family Scombridae, for example, which has dozens of mitogenomes deposited in the NCBI. In this way, our work aimed to assemble six new mitochondrial genomes for the group, expand the knowledge of the evolution and mitochondrial diversity of the family and provide new perspectives on their phylogenetic relationships.
We obtained raw library data of Chiasmodon niger (SRX10444742), Dysalotus oligoscolus (SRX7174474), Kali macrodon (SRX7174492), Kali macrura (SRX7174493), Kali normani (SRX7174494), and Pseudoscopelus astronesthidens (SRX7174526) from the Sequence Reads Archive (SRA) NCBI (Table 1).
While the raw data from C. niger were obtained by targeted enrichment of single copy exons (exon capture) [7], the other data were obtained by target capture of ultraconserved nuclear elements (UCEs) [12]. Although both types of sequencing do not include complete genome sequences, sequence capture is not 100% accurate, and due to the presence of off-target sequences, it is possible to assemble mitogenomes from both exome and UCEs data [13,14].
We imported the raw data into the Galaxy Europe platform [15] and used NOVOplasty v4.3.1 [16] to assemble the mitochondrial genomes by the "de novo" method, ie without reference. Following the instructions of the NOVOplasty software (available at: https://github.com/ndierckx/NOVOPlasty) we did not filter or quality trim the reads and used the raw genome dataset. As a seed, we used the complete mitogenome of K. indica (NC_022488.1) in the K. macrodon and K. normani assembling, and for the other species, we used the corresponding COI mitochondrial gene sequences deposited in GenBank (Supplementary Material 1).
We annotated all mitogenomes using MitoAnnotator [17] available on the MitoFish server (http://mitofish.aori.u-tokyo.ac.jp). We used the BLAST Ring Image Generator (BRIG) [18] to perform a comparative BLAST analysis of available Chiasmodontidae mitogenomes (literature and our assemblies) against our C. niger mitogenome assembly. We used the Tandem Repeats Finder [19] to visualize possible tandem repeat sequences in the mitogenomes.
To perform the phylogenetic analyses, we manually extracted the sequences of the 13 protein-coding genes (PCGs) from our assemblies and added to the dataset the same genes from the mitogenomes of K. indica (NC_022488.1), D. alcoki (NC_022482.1), C. harteli (AP012975.1), 42 mitogenomes from the other 15 families of Scombriformes (Supplementary Material 2), and Aeoliscus strigatus (Syngnantiformes) as an outgroup (NC_010270.1). We aligned the sequences with Muscle [20] in Mega X software [21] and concatenated the alignments in SequenceMatrix v1.8 [22]. We constructed the phylogeny by the maximum likelihood method in IqTree v2.1.2 software [23] using 1,000 ultrafast bootstrap replications and using the evolutionary model TVM+F+R6 estimated by the ModelFinder [24] of IqTree.
The mitochondrial genomes have been deposited in GenBank under the following accession numbers: C. niger, ON831389; D. oligoscolus, ON831390; K. macrodon, ON831391; K. macrura, ON831392; K. normani, ON831393 and P. astronesthidens, ON831394.
As expected, all mitogenomes showed the same arrangement and gene content (Fig. 1), with 13 PCGs, 22 tRNAs, two rRNAs, and a control region (D-loop), with all PCGs (except ND6) in the heavy chain and eight tRNAs in the light chain, as has been observed in most vertebrate mitogenomes, including teleosts, described [25].
We deposited all mitogenomes in GenBank (Supplementary Material 1). Mitogenomes ranged from 16,468 bp in P. astronesthidens to 16,627 bp in K. macrodon and K. normani. We found tandem repeats in the D-loop of all three species of the genus Kali, with three repeating sequences in K. macrura and five in K. macrodon and K. normani (Supplementary Material 3), but we did not find repeat regions in the mitogenomes of C. niger, D. oligoscolus, and P. astronesthidens. Repetitive sequences in the D-loop shared between species of the same genus have already been reported for fishes from the tribe Gymnocharacini and may play a key role in the study of the evolutionary history of these groups [26].
Among the intrafamilial phylogenetic relationships observed for Chiasmodontidae, all species of the genera Chiasmodon and Kali formed monophyletic groups, and the two Dysalotus species used did not cluster (Fig. 2), rather than observed in previous phylogenomic studies [8]. Chiasmodontidae formed a clade with Tetragonuridae and Amarsipidae, corroborating previous phylogenomics and mitogenomics studies [4,7,8,12]. As observed in previous studies [4,6-8,12], other interfamilial phylogenetic relationships recovered in this work were: Stromateidae as a sister group of Nomeidae + Ariommatidae and Trichiuridae as a sister group of Gempylidae (Fig. 2). These clusters correspond to clades A (Stromateidae, Ariommatidae and Nomeidae), C (Chiasmodontidae, Tetragonuridae and Amarsipidae) and partially B (Gempylidae and Trichiuridae, but not Scombrolabracidae) described by Arcila et al. [7].
In the same way Miya et al. [4] and Campbell et al. [6], but unlike Friedman et al. [12], Arcila et al. [7], and Harrington et al. [8], we recovered Caristiidae as a sister group to Icosteidae and Pomatomidae as a sister group to Arripidae. As well as the mitogenomic analysis by Miya et al. [4] we also recovered Gempylidae as a paraphyletic group related to Trichiuridae, and Centrolophidae as a sister group of Scombrolabracidae (Fig. 2). However, the relationships between larger clades containing two or more closely related families remain uncertain and discordant among the different phylogenetic analyzes cited here.
The increase in the number of published mitogenomes associated with the stable organization of this genome among vertebrates, the improvement of assembly techniques, and its maternal nature without recombination make the mitochondrial genome a tool with great potential to solve taxonomic problems and phylogenetic relationships [26,27]. Furthermore, as all mitochondrial genes are linked on the same chromosome, they have a very similar phylogenetic signal and share a unique phylogenetic history, allowing them to be used concatenated and partitioned in phylogenetic analyses [28]. However, incongruities between nuclear and mitochondrial data are frequently reported in the literature. These inconsistencies may be related to data biases and limitations such as saturation, incomplete lineage sampling, differences in taxa sampling, gene partitioning, and other phylogenetic methods employed [27,29-31]. Important biological factors may underlie these divergences, such as past events of hybridizations, selection, and complex biogeographic events with secondary contact of allopatric populations and replacement of mitochondria [29,31,32].
Since the Scombriformes experienced rapid adaptive radiation after the Cretaceous-Paleogene mass extinction [4,12] the high degree of uncertainty reported in the phylogenetic relationships of the group is natural [6,7]. Although some interfamilial phylogenetic relationships, such as Chiasmodontidae with Tetragonuridae and Amarsipidae, are strongly supported, major clade relationships remain uncertain even when phylogenomic approaches are employed, which may result from complex biological events during post-mass extinction adaptive irradiation, as hybrids between lineages, like the radiation of placental mammals [33]. In this way, the phylogenetic representation through two-dimensional trees may not be the best way to illustrate the evolutionary history of the Scombriformes, by “hiding” such events. Finally, we strongly suggest that future phylogenetic analyzes of the group not only increase the amount of data used but also incorporate phylogenetic network analyses. Compared to conventional phylogenetic trees, phylogenetic networks can visualize in a single image conflict between different phylogenetic hypotheses using crosslinks between branches. This type of analysis can be used when complex biological events, called reticulated events, such as hybridizations, recombination, horizontal gene transfer, duplications or gene loss are suspected [33,34].
Notes
Acknowledgments
This study was financed in part by scholarship grants from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil - CAPES (www.capes.gov.br/) (awarded to IHRO).
Supplementary Materials
Supplementary Table is available from the Figshare repository (https://doi.org/10.6084/m9.figshare.19952939). Raw sequencing reads are available at NCBI Sequence Read Archive (SRA), under BioProject number PRJNA561597 and PRJNA644198.
Fig. 1.
Comparative mitogenomics analysis of all the nine chiasmodontid fishes against a reference (Chiasmodon niger), generated by BLAST Ring Image Generator (BRIG). Gaps in rings correspond to regions with <50% identity to the reference sequence (BLAST comparison).
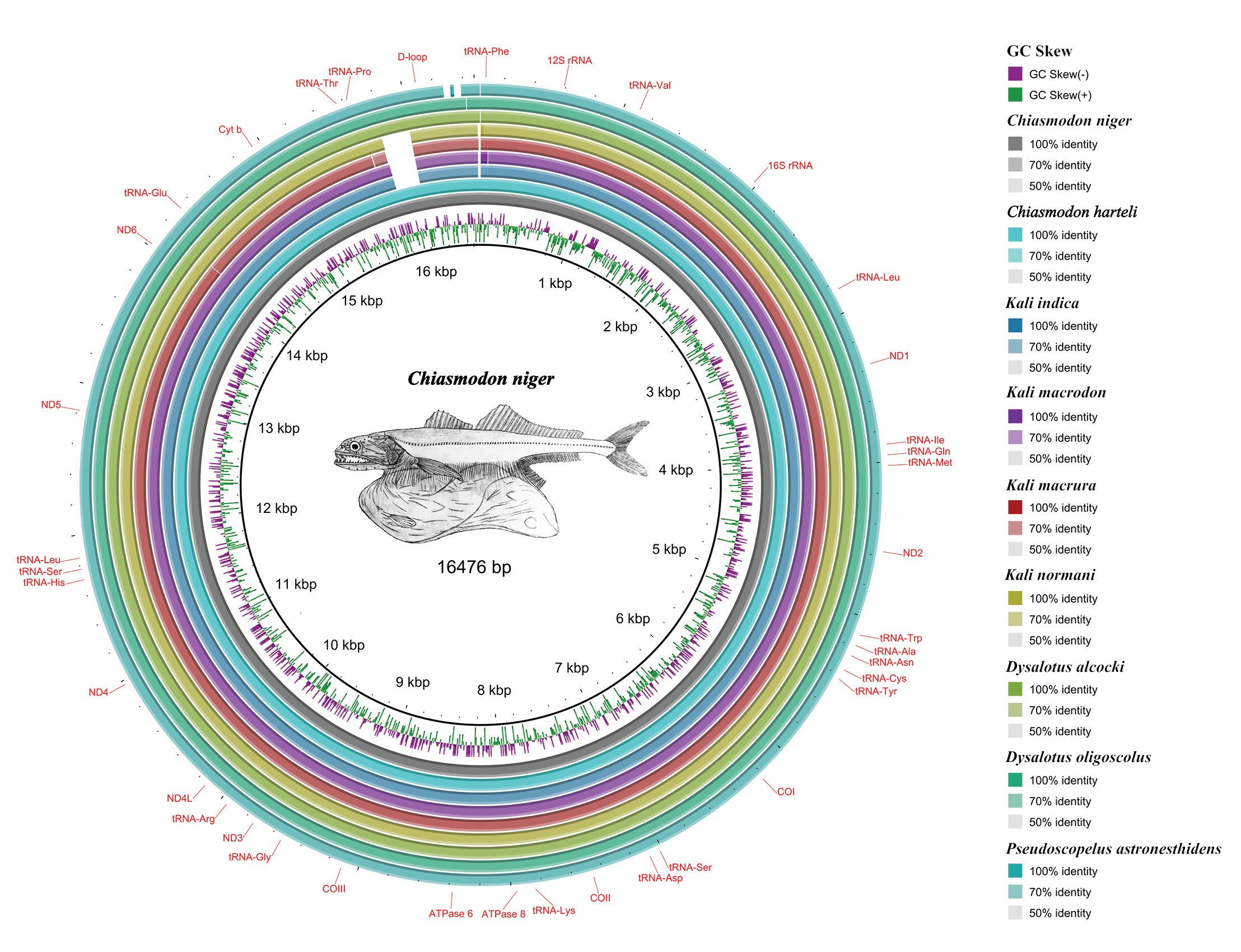
Fig. 2.
The phylogenetic tree of Chiasmodontidae mitogenomes and other 43 species available in GenBank. The bootstrap values were indicated in each branch of the tree. Aeoliscus strigatus was selected as an outgroup.
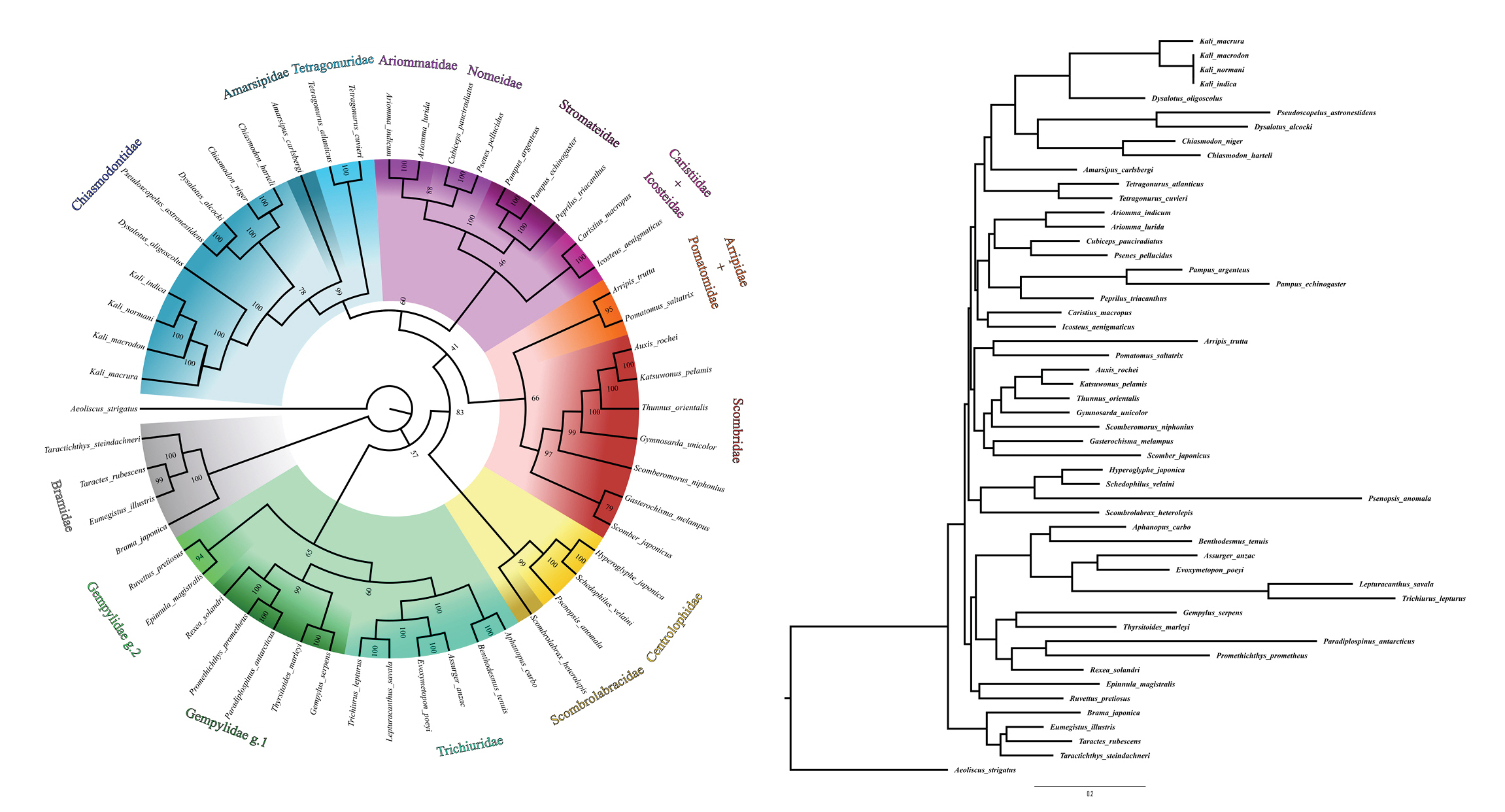
Table 1.
Raw genomic data obtained from the NCBI for each species
References
1. Fricke R, Eschmeyer WN, Fong JD. Genera/species by family/subfamily in Eschmeyer’s Catalog of Fishes. San Francisco: California Academy of Sciences, Accessed 2022 Jun 24. Available from: https://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp.
2. Marshall NB. Developments in Deep-Sea Biology. Poole: Blandford Press, 1979.
3. Melo MR. Revision of the genus Chiasmodon (Acanthomorpha: Chiasmodontidae), with the description of two new species. Copeia 2009;2009:583–608.

4. Miya M, Friedman M, Satoh TP, Takeshima H, Sado T, Iwasaki W, et al. Evolutionary origin of the Scombridae (tunas and mackerels): members of a paleogene adaptive radiation with 14 other pelagic fish families. PLoS One 2013;8:e73535.



5. Betancur RR, Wiley EO, Arratia G, Acero A, Bailly N, Miya M, et al. Phylogenetic classification of bony fishes. BMC Evol Biol 2017;17:162.




6. Campbell MA, Sado T, Shinzato C, Koyanagi R, Okamoto M, Miya M. Multilocus phylogenetic analysis of the first molecular data from the rare and monotypic Amarsipidae places the family within the Pelagia and highlights limitations of existing data sets in resolving pelagian interrelationships. Mol Phylogenet Evol 2018;124:172–180.


7. Arcila D, Hughes LC, Melendez-Vazquez B, Baldwin CC, White WT, Carpenter KE, et al. Testing the utility of alternative metrics of branch support to address the ancient evolutionary radiation of tunas, stromateoids, and allies (Teleostei: Pelagiaria). Syst Biol 2021;70:1123–1144.



8. Harrington RC, Friedman M, Miya M, Near TJ, Campbell MA. Phylogenomic resolution of the monotypic and enigmatic Amarsipus, the bagless glassfish (Teleostei, Amarsipidae). Zool Scr 2021;50:411–422.


9. Nelson JS, Grande TC, Wilson MV. Fishes of the World. 5th ed. Hoboken: John Wiley & Sons, 2016.
10. Orrell TM, Collette BB, Johnson GD. Molecular data support separate scombroid and xiphioid clades. Bull Mar Sci 2006;79:505–519.
11. Near TJ, Dornburg A, Eytan RI, Keck BP, Smith WL, Kuhn KL, et al. Phylogeny and tempo of diversification in the superradiation of spiny-rayed fishes. Proc Natl Acad Sci U S A 2013;110:12738–12743.



12. Friedman M, Feilich KL, Beckett HT, Alfaro ME, Faircloth BC, Cerny D, et al. A phylogenomic framework for pelagiarian fishes (Acanthomorpha: Percomorpha) highlights mosaic radiation in the open ocean. Proc Biol Sci 2019;286:20191502.




13. Mobili F, Miyaki CY, Pellegrino KC, et al. Ultraconserved elements sequencing as a low-cost source of complete mitochondrial genomes and microsatellite markers in non-model amniotes. PLoS One 2015;10:e0138446.



14. Picardi E, Pesole G. Mitochondrial genomes gleaned from human whole-exome sequencing. Nat Methods 2012;9:523–524.



15. Jalili V, Afgan E, Gu Q, Clements D, Blankenberg D, Goecks J, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res 2020;48:W395–W402.




16. Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res 2017;45:e18.


17. Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, et al. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol 2013;30:2531–2540.



18. Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 2011;12:402.




19. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 1999;27:573–580.



20. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004;32:1792–1797.



21. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 2018;35:1547–1549.



22. Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011;27:171–180.


23. Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 2020;37:1530–1534.



24. Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 2017;14:587–589.




25. Miya M, Kawaguchi A, Nishida M. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol 2001;18:1993–2009.


26. Pasa R, Menegidio FB, Rodrigues-Oliveira IH, da Silva IB, de Campos ML, Rocha-Reis DA, et al. Ten complete mitochondrial genomes of Gymnocharacini (Stethaprioninae, Characiformes). insights into evolutionary relationships and a repetitive element in the control region (D-loop). Front Ecol Evol 2021;9:650783.

27. Tamashiro RA, White ND, Braun MJ, Faircloth BC, Braun EL, Kimball RT. What are the roles of taxon sampling and model fit in tests of cyto-nuclear discordance using avian mitogenomic data? Mol Phylogenet Evol 2019;130:132–142.


28. Brandley MC, Schmitz A, Reeder TW. Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Syst Biol 2005;54:373–390.


29. Crespi BJ, Fulton MJ. Molecular systematics of Salmonidae: combined nuclear data yields a robust phylogeny. Mol Phylogenet Evol 2004;31:658–679.


30. De Re FC, Robe LJ, Wallau GL, Loreto EL. Inferring the phylogenetic position of the Drosophila flavopilosa group: incongruence within and between mitochondrial and nuclear multilocus datasets. J Zool Syst Evol Res 2017;55:208–221.


31. Zadra N, Rizzoli A, Rota-Stabelli O. Chronological incongruences between mitochondrial and nuclear phylogenies of Aedes mosquitoes. Life (Basel) 2021;11:181.



32. Hawkins MT, Leonard JA, Helgen KM, McDonough MM, Rockwood LL, Maldonado JE. Evolutionary history of endemic Sulawesi squirrels constructed from UCEs and mitogenomes sequenced from museum specimens. BMC Evol Biol 2016;16:80.




- TOOLS
- Related articles in GNI








