2. Merlo-Pich E, Alexander RC, Fava M, Gomeni R. A new population-enrichment strategy to improve efficiency of placebo-controlled clinical trials of antidepressant drugs. Clin Pharmacol Ther 2010;88:634ŌĆō642.


4. Hollingsworth SJ. Precision medicine in oncology drug development: a pharma perspective. Drug Discov Today 2015;20:1455ŌĆō1463.


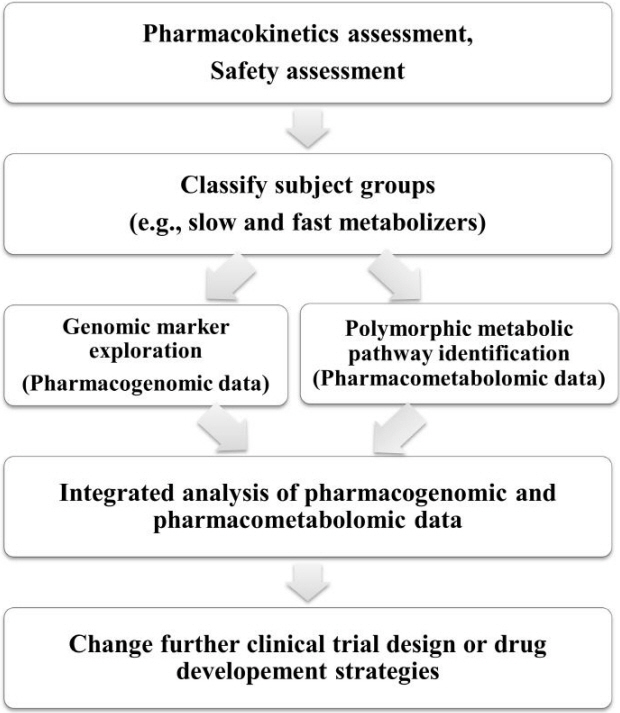
6. Ahn C. Pharmacogenomics in drug discovery and development. Genomics Inform 2007;5:41ŌĆō45.
8. Liou SY, Stringer F, Hirayama M. The impact of pharmacogenomics research on drug development. Drug Metab Pharmacokinet 2012;27:2ŌĆō8.


10. Robertson DG, Frevert U. Metabolomics in drug discovery and development. Clin Pharmacol Ther 2013;94:559ŌĆō561.


11. Kaddurah-Daouk R, Weinshilboum RM, Pharmacometabolomics Research Network. Pharmacometabolomics: implications for clinical pharmacology and systems pharmacology. Clin Pharmacol Ther 2014;95:154ŌĆō167.


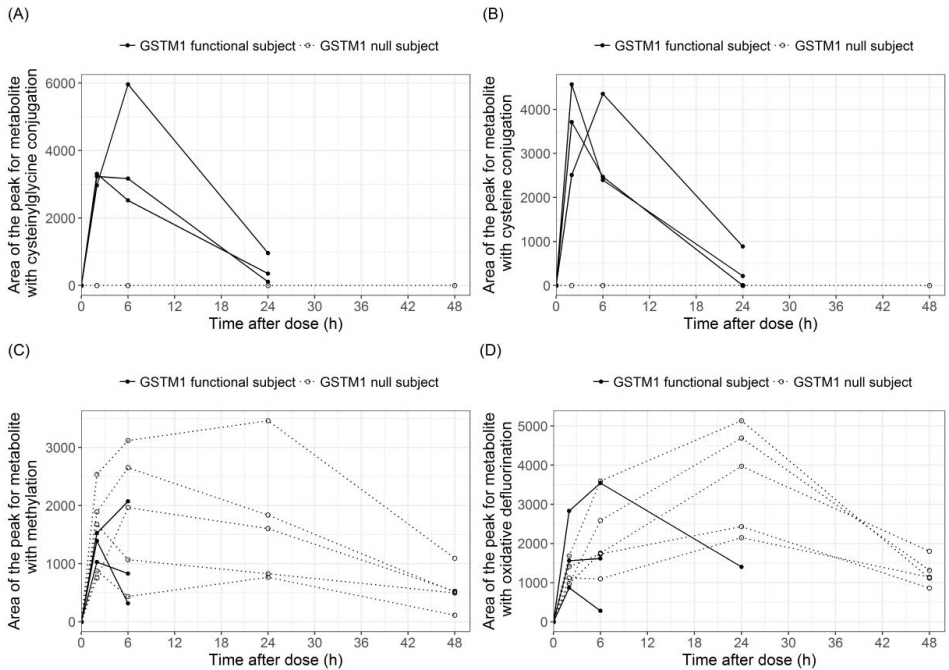
12. Shin KH, Choi MH, Lim KS, Yu KS, Jang IJ, Cho JY. Evaluation of endogenous metabolic markers of hepatic CYP3A activity using metabolic profiling and midazolam clearance. Clin Pharmacol Ther 2013;94:601ŌĆō609.


13. Kim B, Lee J, Shin KH, Lee S, Yu KS, Jang IJ,
et al. Identification of omega- or (omega-1)-hydroxylated medium-chain acylcarnitines as novel urinary biomarkers for CYP3A activity. Clin Pharmacol Ther 2018;103:879ŌĆō887.


14. Yi S, An H, Lee H, Lee S, Ieiri I, Lee Y,
et al. Korean, Japanese, and Chinese populations featured similar genes encoding drug-metabolizing enzymes and transporters: a DMET Plus microarray assessment. Pharmacogenet Genomics 2014;24:477ŌĆō485.


15. Fernandez CA, Smith C, Yang W, Lorier R, Crews KR, Kornegay N,
et al. Concordance of DMET plus genotyping results with those of orthogonal genotyping methods. Clin Pharmacol Ther 2012;92:360ŌĆō365.



17. Rowbotham MC, Nothaft W, Duan WR, Wang Y, Faltynek C, McGaraughty S,
et al. Oral and cutaneous thermosensory profile of selective TRPV1 inhibition by ABT-102 in a randomized healthy volunteer trial. Pain 2011;152:1192ŌĆō1200.


18. Quiding H, Jonzon B, Svensson O, Webster L, Reimfelt A, Karin A,
et al. TRPV1 antagonistic analgesic effect: a randomized study of AZD1386 in pain after third molar extraction. Pain 2013;154:808ŌĆō812.


20. Kurose K, Sugiyama E, Saito Y. Population differences in major functional polymorphisms of pharmacokinetics/pharmacodynamics-related genes in Eastern Asians and Europeans: implications in the clinical trials for novel drug development. Drug Metab Pharmacokinet 2012;27:9ŌĆō54.


22. Fondi M, Lio P. Multi-omics and metabolic modelling pipelines: challenges and tools for systems microbiology. Microbiol Res 2015;171:52ŌĆō64.


23. Kamoun A, Idbaih A, Dehais C, Elarouci N, Carpentier C, Letouze E,
et al. Integrated multi-omics analysis of oligodendroglial tumours identifies three subgroups of 1p/19q co-deleted gliomas. Nat Commun 2016;7:11263.