 |
 |
- Search
| Genomics Inform > Volume 14(3); 2016 > Article |
Abstract
Solid tumor is generally observed in tissues of epithelial or endothelial cells of lung, breast, prostate, pancreases, colorectal, stomach, and bladder, where several genes transcription is regulated by the microRNAs (miRNAs). Argonaute (AGO) protein is a family of protein which assists in miRNAs to bind with mRNAs of the target genes. Hence, study of the binding mechanism between AGO protein and miRNAs, and also with miRNAs-mRNAs duplex is crucial for understanding the RNA silencing mechanism. In the current work, 64 genes and 23 miRNAs have been selected from literatures, whose deregulation is well established in seven types of solid cancer like lung, breast, prostate, pancreases, colorectal, stomach, and bladder cancer. In silico study reveals, miRNAs namely, miR-106a, miR-21, and miR-29b-2 have a strong binding affinity towards PTEN, TGFBR2, and VEGFA genes, respectively, suggested as important factors in RNA silencing mechanism. Furthermore, interaction between AGO protein (PDB ID-3F73, chain A) with selected miRNAs and with miRNAs-mRNAs duplex were studied computationally to understand their binding at molecular level. The residual interaction and hydrogen bonding are inspected in Discovery Studio 3.5 suites. The current investigation throws light on understanding miRNAs based gene silencing mechanism in solid cancer.
Cancer has a complex pathophysiology unlike other diseases. The development of cancer is attributed to a number of predisposing factors like familial and genetic factors, racial and geographic factors and various epidemiological factors like smoking, use of tobacco, diet, and certain type of infection. Majority of the cancers occur in ageing population [1], however there is a changing trend towards younger population in recent years. The uncontrolled division of cancer cells generally spreads to the other part of body through the blood and lymphatic systems. In this context, solid cancer which is an abnormal mass of tissue that usually does not contain cysts or liquid area can be either benign or malignant in nature. Such solid cancers are observed in tissues of epithelial or endothelial cells of lung, breast, prostate, pancreatic, colorectal, stomach, and bladder. Management of cancer is a multimodality approach and includes surgery, radiotherapy, chemotherapy and targeted therapy with certain limitations of each approach. Research is going across the globe for the management of cancer. Recently microRNAs (miRNAs) have drawn attention of the researchers to discover new insights into the treatment of cancer. These miRNAs are highly conserved non-coding single stranded RNAs (~18 to 22 bp) that target mRNA at 3' un-translated region of protein coding genes and act as natural inhibitor. These miRNAs participate in several biological processes such as cell differentiation, proliferation, metabolism, and apoptosis and are also involved in the regulation of gene expression [2]. However, in development of pathologies like cancer, deregulation of miRNAs expression has been identified as potential area to be studied. The miRNAs have special signature and variation from normal cells [3]. Hence, use of specific miRNAs as biomarker may be applied for cancer diagnosis and treatment. Since the expression of tumor suppressor genes and oncogenes are influenced by the miRNAs, provides scope to study their association in cancer [3]. Collating data for miRNAs signatures associated with solid tumors, it was reported that more than 50% of miRNAs genes are located in cancer associated genomic regions or in fragile sites, suggesting that miRNAs play a crucial role in the pathogenesis of a limited range of human cancers than previously expected [4]. Considering their influence on the cancer cell phenotype, some miRNAs are considered to be oncogenic, and others to be tumor-suppressive [5]. Few miRNAs are well characterized in cancer association such as miR-21, miR-17-5p, miR-191, miR-29b-2, miR-223, miR-128b, miR-199a-1, miR-24-1, miR-24-2, miR-146, miR-155, miR-181b-1, miR-20a, miR-107, miR-32, miR-92-2, miR-214, miR-30c, miR-25, miR-221, miR-106a, miR-126, and miR-31 (Supplementary Table 1) that are involved in either up regulation or down regulation of many genes [42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58]. Till date various researchers have explored the unusual expression of miRNAs, and its prognostic significance in various human malignancies. Hence, to study the relationship between miRNAs and cancer, it is important to examine the expression profile of miRNAs and their target genes in different cancer types. As a case study, the unusual expression of miRNA-21 has been conformed in various cancers such as lung, breast, prostate, colorectal, pancreatic, bladder, and stomach cancer, and interestingly the oncogenic like function of miRNA-21 is mediated through the expression of its target genes like PDCD4, PTEN, and TGFBR2 which are generally tumor suppressor genes in nature [59]. Though miRNAs control the gene expression to some extent during translation process but the translational suppression mechanism is still controversial, for example some miRNAs down-regulate while others up-regulate the gene expression. Hence, study of interaction mechanism between miRNAs and their target genes is essential. In the process of miRNAs mediated gene regulation, the Argonaute (AGO) class of proteins play a significant role in the hybrid formation between miRNA-mRNA [60,61]. Despite of several researches, controlling this abnormal cell proliferation selectively in different cancer by miRNAs as natural inhibitor is still a major challenge. In the present study, attempts have been made to develop a strategy for identification and selection of suitable miRNAs, their target genes associated in solid tumor, and as well as to study the role of AGO protein in their interaction at molecular level using computational tools and techniques.
Extensive literature search and exploration of online databases were conducted to understand miRNAs mediated regulation of solid cancer associated target genes using miRCancer (http://mircancer.ecu.edu/) and miRDisease (http://mir2disease.org). miRNAs and target genes were screened by following two criteria: (1) only up regulated oncogenes or down regulated tumor suppressor genes were selected and (2) the screening was performed by selecting oncogenes and tumor suppressor genes as target genes when both of them are either up regulated or down regulated. Further, minimum free energy (MFE) score based binding affinity between miRNAs and their target genes were explored and inspected using miRTarbase web server (http://mirtarbase.mbc.nctu.edu.tw/). Furthermore, the involvement of target genes in different biological function was inspected using UniProt (http://www.uniprot.org/) web server and plotted using Venn (http://bioinformatics.psb.ugent.be/webtools/Venn/) diagram.
To verify the folding affinity between selected miRNAs and their target genes, secondary structures were predicted using RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). The resulted dot bracketed structures of duplex were used for prediction of their tertiary structure using RNA COMPOSER (http://rnacomposer.cs.put poznan.pl/).
The three-dimensional structure of AGO protein was retrieved from Protein Data Bank (PDB ID: 3F73). The structure preparation and correction of AGO protein were performed using Discovery Studio 3.5 suite (http://accelrys.com/products/discovery-studio/visualizatation-download.php).
Docking between three dimensional structure of protein and ligand, may be a biomolecule like miRNA or a gene is an efficient computational method to inspect the molecular interaction [61]. Docking were performed between AGO protein and miRNA-mRNA duplexes (miR-106 and PTEN; miR-21 and TGFBR2; and miR-29b-2 and VEGFA) using PatchDock web server (http://bioinfo3d.cs.tau.ac.il/PatchDock/),algorithm ranked the docked complexes on the basis of highest geometrical shape complementary scores. The molecular interaction was studied using Discovery Studio 3.5 suite.
From the literature, 23 miRNAs were found to be involved in seven solid tumors namely; lung, breast, colorectal, pancreatic, prostate, stomach, and bladder [62]. Again through exploration of online databases 64 different target genes (oncogenes and tumor suppressor genes) were observed to be regulated by these 23 miRNAs (Supplementary Table 1). Again out of these seventeen miRNAs are suspected to be effectively involved in controlling the expression pattern of 45 target genes associated in seven solid tumors (Table 1) [6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41]. Further, binding affinity was explored using miRTarbase web server. Fifteen miRNAs and 23 target genes were selected as per the availability of MFE score (Supplementary Table 2) and subjected to further study. The involvement of these 23 genes in different biological functions are explored and inspected from UniProt web server (http://www.uniprot.org/). It was observed only PTEN, TGFBR2, and VEGFA genes mostly regulating angiogenesis, apoptosis, cell cycle, cell proliferation, and other biological functions (Table 2, Fig. 1). Further, the conservation in binding pattern among selected miRNAs and mRNAs (Table 3) were studied through sequence similarity algorithm using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) web server. The study revealed for the existence of quite less portion of conservation at sequence level among chosen 23 mRNAs and also in 15 miRNAs (Fig. 2), is not sufficient to throw light on binding patterns. Therefore, the binding affinity was evaluated between mRNA and miRNA at secondary structural level. Good MFE score suggested, the target genes PTEN, TGFBR2, and VEGFA are having a strong affinity towards miR-106a, miR-21, and miR-29b-2 (Table 3). Hence those genes and miRNAs were taken into consideration to study their interaction at molecular level.
Duplex sequences between miR-106 and PTEN, miR-21 and TGFBR2, miR-29b-2 and VEGFA were extracted from miRTarbase web server and prediction of secondary folding pattern was performed in RNA fold. The secondary folding patterns along with their binding energy value are reported (Table 4). The predicted binding energy suggested for a high affinity among selected miRNA-mRNA duplexes. Predicted three dimensional models [63] of the duplex structures between miR-106 and PTEN, miR-21 and TGFBR2, miR-29b-2 and VEGFA also supported for strong molecular interaction between them.
The three-dimensional crystallized structure of AGO protein, with 685 amino acid residues was extracted from Protein Data bank (PDB ID: 3F73). The structure of AGO protein contains guide DNA and target RNA duplexes. All water molecules and ligands were removed from the structure. As, the AGO protein is a homo dimer, out of two chains, only chain 'A' of AGO protein was considered and refined before docking. The necessary correction in bond order and bond length of all atoms in the structure was performed using prepare protein and clean geometry protocol of Discovery Studio 3.5.
The AGO protein is a key player in the formation of the RNA-induced silencing complex, a major component of RNA interference. The three dimensional structures of miRNA-mRNA duplexes were prepared using RNA COMPOSER web server (Fig. 3). The first round of docking was performed between the miRNAs and AGO (PDB ID: 3F73, chain A) protein to inspect the binding affinity between the complexes. Out of 10 different poses resulted for docking complexes in Patch Dock server, the pose with highest geometrical shape complementary score [64] is considered as the best docked complex. The resulted geometrical shape complementary score and atomic contact energy scores are reported (Table 5), implicated a strong binding affinity between the miRNAs and AGO protein. The binding affinity between the AGO protein and miRNAs (miR-106a, miR-21, and miR-29b-2) is established through the observation of amino acids (miR-106a: LEU 132, ALA 133, VAL 152, LEU 153, ALA 170, ILE 173, LEU 267, LEU 279, ALA 479,VAL 620, VAL 663, VAL 666, and ILE 671; miR-21: ILE 173, VAL 264, LEU 267, LEU 279, ALA 354, ALA 414, ILE 434, ALA 644, ALA 648, and VAL 685; miR-29b-2: VAL 152, LEU 153, ALA 170, LEU 279, ALA 414, ALA 479, LEU 596, VAL 620, ALA 648, LEU 652, ALA 659, LEU 662, and VAL 663) which are strongly hydrophobic in nature and also amino acids with aromatic rings (miR-106a: TYR 135, TYR 171, TRP415, and PHE 487; miR-21: TYR 135, TYR 171, TRP 415, TYR 642, PHE 647, and PHE 649; miR-29b-2: TRP 156, TRP 415, TYR 642, PHE 647, and PHE649) within a distance of 3.5Å (Table 6, Fig. 4). The presence of hydrogen bonding pattern during interaction (Table 7, Fig. 5) also supported the fact of AGO protein driven miRNA based gene regulation.
The second round docking was performed between AGO protein and miRNA-mRNA duplexes (miR-106 and PTEN; miR-21 and TGFBR2; and miR-29b-2 and VEGFA) separately in PatchDock web server, and scores are reported (Table 8). Amino acids which are strong hydrophobic in nature and amino acids with aromatic rings which are relatively hydrophobic generally contribute a lot towards the stability of binding during molecular interaction between two macro molecules. Hence the molecular interaction between miRNA-mRNA duplexes and AGO protein was studied by inspecting close amino acid residues of AGO protein within a distant of 3.5 Å. Strong hydrophobic amino acids of AGO like VAL 152, LEU 153, VAL 264, LEU 265, LEU 267, LEU 277, ALA 278, LEU 279, ALA 414, ILE 434, ALA 479, VAL 663, and VAL 685 in miR-106a and PTEN duplex; LEU 45, LEU 46, ALA 47, VAL 49, ALA 50, ALA 80, ILE 173, LEU 265, LEU 267, LEU 277, ALA 278, LEU 279, LEU 281, ALA 479, ALA 644, ALA 648, LEU 652, and VAL 663 in miR-21 and TGFBR2 duplex; and ALA 80, LEU 132, VAL 152, LEU 153, ALA 170, ILE 173, VAL 264, ALA 479, VAL 606, ALA 644, ALA 648, VAL 663, and VAL 685 in miR-29b-2 and VEGFA duplex are observed within a distant of 3.5 Å. Similarly, amino acids like TYR 43, TRP 415, PHE 487, and TYR 642 in miR-106a and PTEN duplex; TYR 43, TRP 202, TRP 415, TYR 642, PHE 647, and PHE 649 in miR-21 and TGFBR2 duplex; and TYR 43, TYR 86, TYR 135, TRP 415, TYR 171, TYR 642, PHE 647, and PHE 649 in miR-29b-2 and VEGFA duplex, with aromatic ring are also found as participating in interaction within the binding pocket of AGO protein around distance of 3.5 Å (Table 9, Fig. 6).
The current study focused on in silico identification of genes and their regulation mediated through miRNAs, involved in seven types of solid tumors of colorectal, pancreatic, breast, stomach, lung, prostate, and bladder. miRNAs have a crucial role in gene silencing mechanism observed through the altered expression of certain miRNA molecules in case of neurodegenerative diseases such as Alzheimer and Parkinson suggesting that the association between mRNA-miRNA-AGO, therefore not cancer specific [16,65]. The association between mRNA-miRNA-AGO pattern has been reported [16,65,66] in RNA based gene silencing mechanism and their association at atomic level has already been studied in case of leukemia [25]. In this context, molecular association between AGO protein, miRNAs and mRNAs of target genes were studied and analyzed in case of seven types of solid tumor. Sixty-four genes and 23 miRNAs were collected from the literature (Supplementary Table 1), out of which 45 genes and seventeen miRNAs were selected on the basis of their expression pattern (Table 1). The affinity in binding modes between 15 miRNAs and 23 genes was verified, basing on the availability of MFE scores in miRTarbase web server (Supplementary Table 2). Further, different biological activities of selected genes were studied using UniProt database (http://www.uniprot.org/) to screen out important genes and their contribution towards seven types of solid tumors. The observation suggested three genes namely, PTEN, TGFBR2, and VEGFA were involved in different biological activities like angiogenesis, apoptosis, cell cycle, and cell proliferation, are the major contributing factor in oncogenesis (Table 2). However, quite less sequence conservation was observed within the binding site of 23 target genes (Fig. 2A). But, quite well sequence similarity was observed between miR-106a, miR-17-5p, and miR-20a (Fig. 2B) suggesting for possibility of regulation of PTEN gene through these three miRNAs (Table 3), associated in causing colorectal cancer. Again, a strong and energetically binding affinity was proved through good MFE scores –14.3 (kcal/mol), –16.2 (kcal/mol), and –15 (kcal/mol) for PTEN and miR-106a; TGFBR2 and miR-21; and VEGFA and miR-29b-2, respectively, suggested for appropriate miRNAs selection for PTEN, TGFBR2, and VEGFA genes (Table 3), supported by the predicted binding energy scores for miRNA-mRNA duplex structures (Table 4). Furthermore, to study the molecular basis of AGO protein driven miRNAs namely, miR-106a, miR-21, and miR-29b-2 having highest binding affinity towards their regulating genes namely, PTEN, TGFBR2, and VEGFA respectively, the molecular docking study was carried out. Generally, presence of amino acids with aliphatic group namely, alanine (ALA), valine (VAL), leucine (LEU), and isoleucine (ILE), are strongly hydrophobic in nature provides stability during molecular interaction between macro molecules, whereas amino acids namely, phenylalanine (PHE), tyrosine (TYR), tryptophan (TRP), are relatively hydrophobic in nature, but the presence of aromatic ring like structure provides a steadiness towards binding stability within the structural moiety of a protein. The observation of amino acids (miR-106a: LEU 132, ALA 133, VAL 152, LEU 153, ALA 170, ILE 173, LEU 267, LEU 279, ALA 479,VAL 620, VAL 663, VAL 666, and ILE 671; miR-21: ILE 173, VAL 264, LEU 267, LEU 279, ALA 354, ALA 414, ILE 434, ALA 644, ALA 648, and VAL 685; miR-29b-2: VAL 152, LEU 153, ALA 170, LEU 279, ALA 414, ALA 479, LEU 596, VAL 620, ALA 648, LEU 652, ALA 659, LEU 662, and VAL 663) which are strongly hydrophobic in nature and amino acids with aromatic rings (miR-106a: TYR 135, TYR 171, TRP 415, and PHE 487; miR-21: TYR 135, TYR 171, TRP 415, TYR 642, PHE 647, and PHE 649; miR-29b-2: TRP 156, TRP 415, TYR 642, PHE 647, and PHE649) during interaction within a distance of 3.5Å (Table 6, Fig. 4) recognized a strong molecular interaction between AGO protein and miR-106a, miR-21, and miR-29b-2. Again, weak interaction like hydrogen bonding has a major contribution to facilitate the stability of molecules during interaction at atomic level. Here, the amino acids like, LEU 153, PRO 169, TYR 171, GLU 416, PHE 487, ARG 574, and ARG 615; ARG 172, THR 201, LEU 279, ALA 354, GLN355, PRO 412, MET 413, TRP 415, ARG 418, ASN 436, ARG 615, ALA 644, ARG 661, and ARG 668; LYS 101, ARG 114, ASP 159, PRO 412, ALA 414, ASP 478, LYS 575, ARG 580, ARG 580, GLU 597, ASP 598, ARG 615, ALA 648, ASP 660, and ARG 661 are participated in hydrogen bonding interaction with miR-106a, miR-21, and miR-29b-2 respectively in the structural moiety of AGO protein within a distance of 2.5Å (Table 7, Fig. 5), strongly supports for binding stability during assistance of AGO protein for miRNA based gene regulation. Furthermore, presence of commonly participating strong hydrophobic amino acids namely, VAL 663 and amino acid with aromatic rings namely, TYR 43, TRP 415, and TYR 642 during molecular interaction of AGO protein with miR-106a and PTEN, miR-21 and TGFBR2, and miR-29b-2 and VEGFA duplex within a distance of 3.5Å (Table 9, Fig. 6). Although, the evidence at molecular association between mRNA-miRNA-AGO pattern have already been established computationally [16,61] the current investigation recommending for AGO protein assistance in regulation of PTEN, TGFBR2, and VEGFA genes by miR-106a, miR-21, and miR-29b-2, respectively, associated in seven types of solid tumor. This report would be helpful in understanding miRNA-based gene silencing mechanisms in seven types of solid cancer like colorectal cancer, pancreatic cancer, breast cancer, stomach cancer, lung cancer, prostate cancer, and bladder cancer. Furthermore, extensive computational study may be carried out for better understanding the mechanism of miRNAs based gene regulation in solid cancer. This methodology would further help us to design suitable miRNAs against respective genes rationally, only after validation through laboratory experiments.
Acknowledgments
Authors are thankful to Department of Biotechnology; Govetment of India for providing BIF Centre facility to carry out the current research work. Also authors are grateful to the authorities of Department of Bioinformatics, Centre for Post Graduate studies, Orissa University of agriculture & Technology, Bhubaneswar, Odisha, India for their constant encouragement and allowing for accomplishing the research work.
Supplementary materials
Supplementary data including two tables can be found with this article online at http://www.genominfo.org/src/sm/gni-14-112-s001.pdf
Supplementary Table 1
Twenty-three miRNAs and 64 targeted genes with their expression type in seven types of solid cancer are reported
Supplementary Table 2
MFE score based binding affinity between 15 miRNAs and 23 target genes associated in seven types of solid cancer are reported
References
1. Alison RM. Encyclopedia of Life Sciences: Cancer. London: Nature Publishing Group, 2001.
3. Grammatikakis I, Gorospe M, Abdelmohsen K. Modulation of cancer traits by tumor suppressor microRNAs. Int J Mol Sci 2013;14:1822–1842. PMID: 23325049.



4. Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol 2007;302:1–12. PMID: 16989803.


5. Esquela-Kerscher A, Slack FJ. Oncomirs–microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259–269. PMID: 16557279.


6. Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 2008;27:4373–4379. PMID: 18372920.


7. Xu LF, Wu ZP, Chen Y, Zhu QS, Hamidi S, Navab R. MicroRNA-21 (miR-21) regulates cellular proliferation, invasion, migration, and apoptosis by targeting PTEN, RECK and Bcl-2 in lung squamous carcinoma, Gejiu City, China. PLoS One 2014;9:e103698. PMID: 25084400.



8. Wang P, Zou F, Zhang X, Li H, Dulak A, Tomko RJ Jr, et al. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res 2009;69:8157–8165. PMID: 19826040.



9. Gong AY, Eischeid AN, Xiao J, Zhao J, Chen D, Wang ZY, et al. miR-17-5p targets the p300/CBP-associated factor and modulates androgen receptor transcriptional activity in cultured prostate cancer cells. BMC Cancer 2012;12:492. PMID: 23095762.



10. Fang L, Li H, Wang L, Hu J, Jin T, Wang J, et al. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget 2014;5:2974–2987. PMID: 24912422.



11. Yamada N, Kitamoto S, Yokoyama S, Hamada T, Goto M, Tsutsumida H, et al. Epigenetic regulation of mucin genes in human cancers. Clin Epigenetics 2011;2:85–96. PMID: 22704331.




12. Nagpal N, Kulshreshtha R. miR-191: an emerging player in disease biology. Front Genet 2014;5:99. PMID: 24795757.



13. Wang H, Guan X, Tu Y, Zheng S, Long J, Li S, et al. MicroRNA-29b attenuates non-small cell lung cancer metastasis by targeting matrix metalloproteinase 2 and PTEN. J Exp Clin Cancer Res 2015;34:59. PMID: 26063204.



14. Liu H, Wang B, Lin J, Zhao L. microRNA-29b: an emerging player in human cancer. Asian Pac J Cancer Prev 2014;15:9059–9064. PMID: 25422179.



15. Ru P, Steele R, Newhall P, Phillips NJ, Toth K, Ray RB. miRNA-29b suppresses prostate cancer metastasis by regulating epithelial-mesenchymal transition signaling. Mol Cancer Ther 2012;11:1166–1173. PMID: 22402125.


16. Wang Y, Li Y, Ma Z, Yang W, Ai C. Mechanism of microRNA-target interaction: molecular dynamics simulations and thermodynamics analysis. PLoS Comput Biol 2010;6:e1000866. PMID: 20686687.



17. Gong J, Li J, Wang Y, Liu C, Jia H, Jiang C, et al. Characterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancer. Carcinogenesis 2014;35:497–506. PMID: 24130168.


18. Wu LH, Cai QQ, Dong YW, Wang R, He BM, Qi B, et al. Decoy oligonucleotide rescues IGF1R expression from microRNA-223 suppression. PLoS One 2013;8:e82167. PMID: 24324762.



19. Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 2008;27:5959–5974. PMID: 18836476.


20. Yin Y, Li J, Chen S, Zhou T, Si J. MicroRNAs as diagnostic biomarkers in gastric cancer. Int J Mol Sci 2012;13:12544–12555. PMID: 23202912.



21. Ding G, Huang G, Liu HD, Liang HX, Ni YF, Ding ZH, et al. MiR-199a suppresses the hypoxia-induced proliferation of non-small cell lung cancer cells through targeting HIF1α. Mol Cell Biochem 2013;384:173–180. PMID: 24022342.


22. Zhang Y, Fan KJ, Sun Q, Chen AZ, Shen WL, Zhao ZH, et al. Functional screening for miRNAs targeting Smad4 identified miR-199a as a negative regulator of TGF-β signalling pathway. Nucleic Acids Res 2012;40:9286–9297. PMID: 22821565.



23. Zhu X, Li H, Long L, Hui L, Chen H, Wang X, et al. miR-126 enhances the sensitivity of non-small cell lung cancer cells to anticancer agents by targeting vascular endothelial growth factor A. Acta Biochim Biophys Sin (Shanghai) 2012;44:519–526. PMID: 22510476.


24. Zhou Y, Feng X, Liu YL, Ye SC, Wang H, Tan WK, et al. Down-regulation of miR-126 is associated with colorectal cancer cells proliferation, migration and invasion by targeting IRS-1 via the AKT and ERK1/2 signaling pathways. PLoS One 2013;8:e81203. PMID: 24312276.



25. Liao YL, Tsai KW, Lin WC. miRNAs in gastric cancer. Rijeka: INTECH Open Access Publisher, 2011.
26. Jia AY, Castillo-Martin M, Bonal DM, Sánchez-Carbayo M, Silva JM, Cordon-Cardo C. MicroRNA-126 inhibits invasion in bladder cancer via regulation of ADAM9. Br J Cancer 2014;110:2945–2954. PMID: 24823697.



27. Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest 2010;120:1298–1309. PMID: 20237410.



28. Zhang Q, Padi SK, Tindall DJ, Guo B. Polycomb protein EZH2 suppresses apoptosis by silencing the proapoptotic miR-31. Cell Death Dis 2014;5:e1486. PMID: 25341040.



29. Lin SL, Chiang A, Chang D, Ying SY. Loss of miR-146a function in hormone-refractory prostate cancer. RNA 2008;14:417–424. PMID: 18174313.



30. Labbaye C, Testa U. The emerging role of miR-146A in the control of hematopoiesis, immune function and cancer. J Hematol Oncol 2012;5:13. PMID: 22453030.



31. Zhang CM, Zhao J, Deng HY. MiR-155 promotes proliferation of human breast cancer MCF-7 cells through targeting tumor protein 53-induced nuclear protein 1. J Biomed Sci 2013;20:79. PMID: 24152184.



32. Liu J, Huang W, Yang H, Luo Y. Expression and function of miR-155 in breast cancer. Biotechnol Biotechnol Equip 2015;29:840–843.

33. Mansueto G, Forzati F, Ferraro A, Pallante P, Bianco M, Esposito F, et al. Identification of a new pathway for tumor progression: microRNA-181b up-regulation and CBX7 down-regulation by HMGA1 protein. Genes Cancer 2010;1:210–224. PMID: 21779448.



34. Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell 2010;39:493–506. PMID: 20797623.



35. Aslam MI, Patel M, Singh B, Jameson JS, Pringle JH. MicroRNA manipulation in colorectal cancer cells: from laboratory to clinical application. J Transl Med 2012;10:128. PMID: 22716183.



36. Lee KH, Lotterman C, Karikari C, Omura N, Feldmann G, Habbe N, et al. Epigenetic silencing of microRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology 2009;9:293–301. PMID: 19407485.



37. Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res 2008;68:6162–6170. PMID: 18676839.



38. Wu W, Yang J, Feng X, Wang H, Ye S, Yang P, et al. MicroRNA-32 (miR-32) regulates phosphatase and tensin homologue (PTEN) expression and promotes growth, migration, and invasion in colorectal carcinoma cells. Mol Cancer 2013;12:30. PMID: 23617834.



39. Zhang XJ, Ye H, Zeng CW, He B, Zhang H, Chen YQ. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J Hematol Oncol 2010;3:46. PMID: 21106054.



40. Azrak SS, Ginel-Picardo A, Drosten M, Barbacid M, Santos E. Reversible, interrelated mRNA and miRNA expression patterns in the transcriptome of Rasless fibroblasts: functional and mechanistic implications. BMC Genomics 2013;14:731. PMID: 24156637.



41. Feng B, Dong TT, Wang LL, Zhou HM, Zhao HC, Dong F, et al. Colorectal cancer migration and invasion initiated by microRNA-106a. PLoS One 2012;7:e43452. PMID: 22912877.



42. Li XM, Wang AM, Zhang J, Yi H. Down-regulation of miR-126 expression in colorectal cancer and its clinical significance. Med Oncol 2011;28:1054–1057. PMID: 20680522.


43. Yu J, Ohuchida K, Mizumoto K, Fujita H, Nakata K, Tanaka M. MicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biol Ther 2010;10:748–757. PMID: 20703102.


44. Wu W, Yang P, Feng X, Wang H, Qiu Y, Tian T, et al. The relationship between and clinical significance of microRNA-32 and phosphatase and tensin homologue expression in colorectal cancer. Genes Chromosomes Cancer 2013;52:1133–1140. PMID: 24123284.


45. Qin S, Zhu Y, Ai F, Li Y, Bai B, Yao W, et al. MicroRNA-191 correlates with poor prognosis of colorectal carcinoma and plays multiple roles by targeting tissue inhibitor of metalloprotease 3. Neoplasma 2014;61:27–34. PMID: 24195505.


46. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257–2261. PMID: 16461460.



47. Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer 2010;126:1166–1176. PMID: 19676045.


48. Baffa R, Fassan M, Volinia S, O'Hara B, Liu CG, Palazzo JP, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol 2009;219:214–221. PMID: 19593777.


49. Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene 2008;27:5643–5647. PMID: 18504431.



50. Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu T, et al. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem 2011;351:157–164. PMID: 21249429.


51. Liu K, Li G, Fan C, Diao Y, Wu B, Li J. Increased expression of microRNA-221 in gastric cancer and its clinical significance. J Int Med Res 2012;40:467–474. PMID: 22613407.


52. Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res 2011;9:824–833. PMID: 21628394.


53. Li ZW, Yang YM, Du LT, Dong Z, Wang LL, Zhang X, et al. Overexpression of miR-223 correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Med Oncol 2014;31:256. PMID: 25270282.


54. Yabushita S, Fukamachi K, Tanaka H, Sumida K, Deguchi Y, Sukata T, et al. Circulating microRNAs in serum of human K-ras oncogene transgenic rats with pancreatic ductal adenocarcinomas. Pancreas 2012;41:1013–1018. PMID: 22513294.


55. Wang Y, Yu Y, Tsuyada A, Ren X, Wu X, Stubblefield K, et al. Transforming growth factor-β regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene 2011;30:1470–1480. PMID: 21102523.



56. Li Y, Vandenboom TG 2nd, Wang Z, Kong D, Ali S, Philip PA, et al. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res 2010;70:1486–1495. PMID: 20124483.



57. Sun Q, Zhao X, Liu X, Wang Y, Huang J, Jiang B, et al. miR-146a functions as a tumor suppressor in prostate cancer by targeting Rac1. Prostate 2014;74:1613–1621. PMID: 25214035.


58. Li BS, Zuo QF, Zhao YL, Xiao B, Zhuang Y, Mao XH, et al. MicroRNA-25 promotes gastric cancer migration, invasion and proliferation by directly targeting transducer of ERBB2, 1 and correlates with poor survival. Oncogene 2015;34:2556–2565. PMID: 25043310.


59. Hamano R, Ishii H, Miyata H, Doki Y, Mori M. Role of microRNAs in solid tumors. J Nucleic Acids Investig 2011;2:e2.

60. Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 2008;9:22–32. PMID: 18073770.


61. Das RP, Konkimalla VB, Rath SN, Hansa J, Jagdeb M. Elucidation of the molecular interaction between miRNAs and the HOXA9 gene, involved in acute myeloid leukemia, by the assistance of Argonaute protein through a computational approach. Genomics Inform 2015;13:45–52. PMID: 26175662.



62. Mascellani N, Tagliavini L, Gamberoni G, Rossi S, Marchesini J, Taccioli C, et al. Using miRNA expression data for the study of human cancer. Minerva Biotecnol 2008;20:23–30.
63. Popenda M, Szachniuk M, Antczak M, Purzycka KJ, Lukasiak P, Bartol N, et al. Automated 3D structure composition for large RNAs. Nucleic Acids Res 2012;40:e112. PMID: 22539264.



64. Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 2005;33:W363–W367. PMID: 15980490.



65. Junn E, Mouradian MM. MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol Ther 2012;133:142–150. PMID: 22008259.



66. Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009;460:479–486. PMID: 19536157.



Fig. 1
The 23 target genes and their involvement in different biological functions like angiogenesis, apoptosis, cell cycle, cell proliferation, and others are plotted using Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/). The numbers denoted in plot indicate the number of target genes involvement in each of the different biological function.

Fig. 2
The sequence similarity between 23 selected genes (A) and 15 miRNAs (B) inspected and deciphered. The sequence conservation in binding pattern highlighted in red box. miRNA, microRNA.

Fig. 3
Three dimensional binding modes of miR-106a and mRNA of PTEN gene (A), miR-21 and mRNA of TGFBR2 gene (B), and miR-29b-2 and mRNA of VEGFA gene (C) are deciphered, respectively.
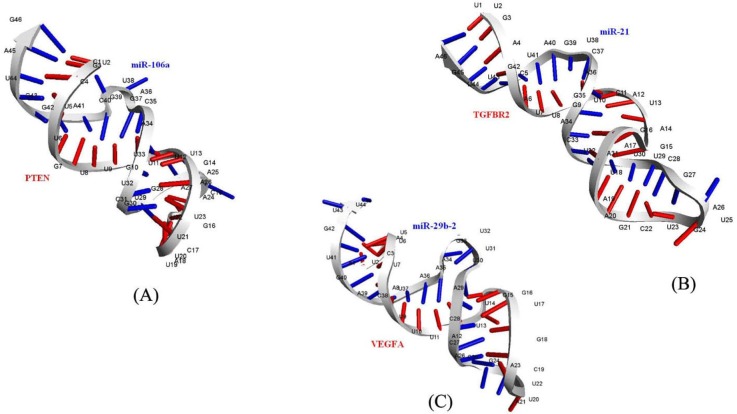
Fig. 4
Amino acid residues of Argonaute protein participating in interaction with miR-106a (A), miR-21 (B), and miR-29b-2 (C) within a distance of 3.5 Å are deciphered, respectively.
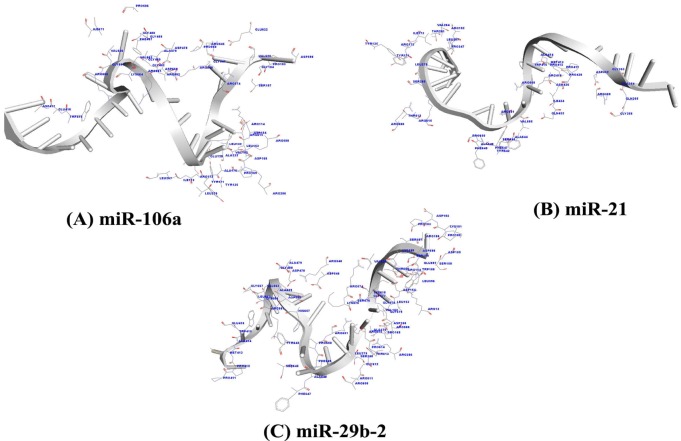
Fig. 5
Amino acid residues of Argonaute protein participating in hydrogen bonding interaction with miR-106a (A), miR-21 (B), and miR-29b-2 (C) within a distance of 2.5 Å are deciphered, respectively.
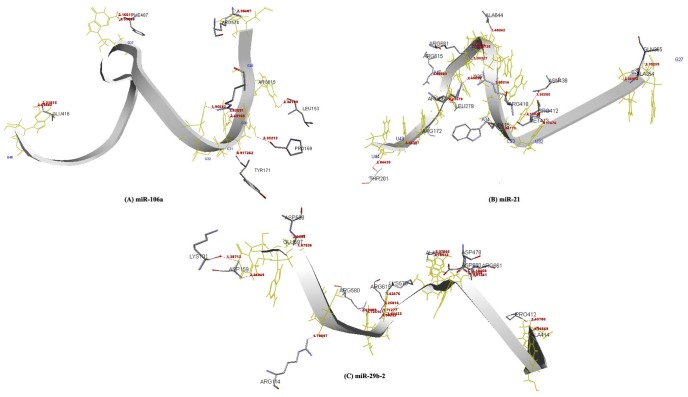
Fig. 6
Amino acid residues of Argonaute protein participating in interaction with miR-106a and mRNA of PTEN gene (A), miR-21 and mRNA of TGFBR2 gene (B), and miR-29b-2 and mRNA of VEGFA gene (C) within a distance of 3.5 Å are deciphered, respectively.
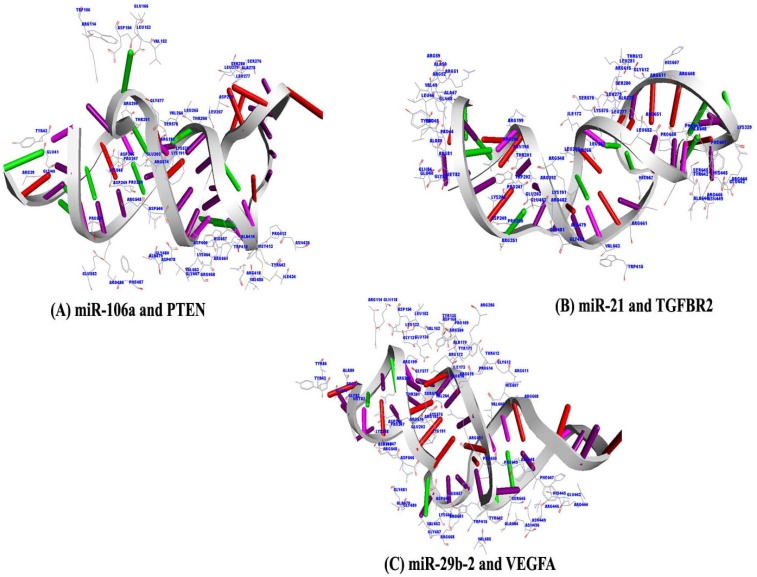
Table 1.
The 17 miRNAs and 45 target genes involved in regulation of seven types of solid cancers are reported
| miRNA | Cancer |
Target gene |
Reference | |||
|---|---|---|---|---|---|---|
| Oncogene | Expression | Tumor suppressor gene | Expression | |||
| miR-21 | Lung | - | - | PDCD4a | Down | [6] |
| Breast | - | - | PTENa | Down | [7] | |
| Colorectal | CDC25Aa | Down | PDCD4a, TGFBR2a | Down | [8] | |
| Bladder | RECKa, BCL2a | Up | Absent | - | [7] | |
| miR-17-5p | Prostate | PCAFa | Up | Absent | - | [9] |
| Colorectal | Absent | - | PTENa | Down | [10] | |
| Pancreatic | MUC17a | Up | Absent | - | [11] | |
| miR-191 | Colorectal | SOX4a, TIMP3a | Up | Absent | - | [12] |
| miR-29b-2 | Lung | MMP2a | Up | Absent | - | [13] |
| Breast | MMP2a, DNMT3Ba | Up | Absent | - | [14] | |
| Prostate | DNMT3Ba, VEGFAa, MMP2a | Up | Absent | - | [15] | |
| Colorectal | MMP2a | Up | Absent | - | [16] | |
| Stomach | MMP2a | Up | Absent | - | [17] | |
| Bladder | IFNGa, MMP2a | Up | Absent | - | [14] | |
| miR-223 | Colorectal | FOXO1a, IGF-1Ra | Up | Absent | - | [18] |
| Pancreatic | NFIAa | Up | Absent | - | [19] | |
| Stomach | Absent | - | FBXW7a | Down | [20] | |
| miR-199a-1 | Lung | HIF1αa | Up | Absent | [21] | |
| Pancreatic | Absent | - | SMAD4a | Down | [22] | |
| miR-126 | Lung | VEGFAa, CRKa, SLC7A5a | Down | EGFRa | Down | [23] |
| Colorectal | Absent | - | IRS-1a | Down | [24] | |
| Stomach | CRKa | Up | Absent | - | [25] | |
| Bladder | ADAM9a | Up | Absent | - | [26] | |
| miR-31 | Lung | Absent | - | LATS2a, PPP2R2Aa | Down | [27] |
| Prostate | EZH2a | Up | Absent | - | [28] | |
| miR-146a | Prostate | ROCK1a | Up | Absent | - | [29] |
| Pancreatic | IRAK1a, NFKBa, MTA2a | Down | EGFRa | Down | [30] | |
| miR-155 | Lung | CASP3a, TP53INP1a, CASP317a | Down | SOCS1a, PTENa | Down | [31] |
| Breast | FOXO3a | Up | SOCS1a | Up | [32] | |
| miR-181b-1 | Breast | BCL2a | Up | Absent | - | [33] |
| Colorectal | TIMP3a | Up | Absent | - | [33] | |
| Stomach | Absent | - | TIMP3a | Down | [34] | |
| miR-20a | Colorectal | Absent | - | PTENa | Down | [35] |
| Pancreatic | ABL2a | Up | Absent | - | [30] | |
| miR-107 | Pancreatic | Absent | - | CDK6a | Down | [36] |
| miR-32 | Prostate | BCL2L11a | Up | Absent | - | [37] |
| Colorectal | Absent | - | PTENa | Down | [38] | |
| mir-214 | Pancreatic | Absent | - | ING4a | Down | [39] |
| miR-106a | Colorectal | E2F1a | Up | PTENa, RB1a | Up | [40] |
| Pancreatic | Absent | - | TGFBR2a | Down | [41] | |
| miR-128b | Lung | VEGFa, CRKa, SLC7A5a | Down | EGFRa | Down | [17] |
Table 2.
Twenty-three target genes and their involvement in different biological function are reported from UniProt (http://www.uniprot.org/) web server
| No. | Gene | miRNA | Function |
|---|---|---|---|
| 1 | TGFBR2a | miR-21, miR-106a | Angiogenesis, apoptosis, cell cycle, cell proliferation, other function |
| 2 | VEGFAa | miR-29b-2, miR-126 | |
| 3 | PTENa | miR-106a, miR-20a, miR-17-5p, miR-32, miR-155, miR-21 | |
| 4 | IFNG | miR-29b-2 | Apoptosis, cell cycle, cell proliferation, other function |
| 5 | CDK6 | miR-107 | |
| 6 | IGF-1R | miR-223 | Apoptosis, cell cycle, other function |
| 7 | TP53INP1 | miR-155 | |
| 8 | MMP2 | miR-29b-2 | Angiogenesis, other function |
| 9 | ADAM9 | miR-126 | |
| 10 | RB1 | miR-106a | Apoptosis, cell cycle |
| 11 | FBXW7 | miR-223 | Apoptosis, other function |
| 12 | EGFR | miR-146a, miR-128b | Cell proliferation, other function |
| 13 | SMAD4 | miR-199a-1 | |
| 14 | CDC25A | miR-21 | Cell proliferation |
| 15 | DNMT3B | miR-29b-2 | Other function |
| 16 | ABL2 | miR-20a | |
| 17 | ROCK1 | miR-146a | |
| 18 | CRK | miR-126 | |
| 19 | PPP2R2A | miR-31 | |
| 20 | MTA2 | miR-146a | |
| 21 | TIMP3 | miR-181b-1 | |
| 22 | RECK | miR-21 | |
| 23 | SLC7A5 | miR-126 |
Table 3.
MFE score based binding affinity between 15 miRNAs and 23 target genes associated in seven types of solid cancer
| No. | Gene | Cancer | miRNA | MFE score (kcal/mol) |
|---|---|---|---|---|
| 1 | PTENa | Colorectal | miR-106a | –14.3 |
| Colorectal | miR-20a | –13 | ||
| Colorectal | miR-17-5p | –12.9 | ||
| Colorectal | miR-32 | –12 | ||
| Lung | miR-155 | –10.7 | ||
| Breast | miR-21 | –9.41 | ||
| 2 | TGFBR2a | Colorectal | miR-21 | –16.2 |
| Pancreatic | miR-106a | –15.1 | ||
| 3 | VEGFAa | Prostate | miR-29b-2 | –15 |
| Lung | miR-126 | –11.5 | ||
| 4 | CDC25A | Colorectal | miR-21 | –10.2 |
| 5 | RECK | Bladder | miR-21 | –16.1 |
| 6 | MMP2 | Lung, breast, colorectal, prostate, stomach, bladder | miR-29b-2 | –11.3 |
| 7 | DNMT3B | Breast, prostate | miR-29b-2 | –12.6 |
| 8 | IFNG | Bladder | miR-29b-2 | –16.7 |
| 9 | IGF-1R | Colorectal | miR-223 | –11 |
| 10 | FBXW7 | Stomach | miR-223 | –12.5 |
| 11 | SMAD4 | Pancreatic | miR-199a-1 | –13.2 |
| 12 | CRK | Lung | miR-126 | –18.1 |
| 13 | SLC7A5 | Lung | miR-126 | –16.3 |
| 14 | ADAM9 | Bladder | miR-126 | –13.8 |
| 15 | PPP2R2A | Lung | miR-31 | –17.9 |
| 16 | ROCK1 | Prostate | miR-146a | –9.3 |
| 17 | MTA2 | Pancreatic | miR-146a | –14.7 |
| 18 | EGFR | Pancreatic | miR-146a | –19.8 |
| Lung | miR-128b | –17.2 | ||
| 19 | TP53INP1 | Lung | miR-155 | –13.4 |
| 20 | TIMP3 | Colorectal, stomach | miR-181b-1 | –14.7 |
| 21 | ABL2 | Pancreatic | miR-20a | –20.4 |
| 22 | CDK6 | Pancreatic | miR-107 | –19.2 |
| 23 | RB1 | Colorectal | miR-106a | –13.7 |
Table 4.
Secondary folding pattern of miRNA-mRNA duplexes along with their predicted binding energy (RNAfold) and MFE scores (miRTarbase)
Table 5.
Docking scores between miRNA and AGO protein
| miRNA and AGO (PDB ID:3F73, chain A) | Score | Area | ACE |
|---|---|---|---|
| miR-106a–AGO | 16,618 | 2,039.00 | –455.23 |
| miR-21–AGO | 16,678 | 2,275.40 | –374.62 |
| miR-29b-2–AGO | 19,222 | 2,942.20 | –555.08 |
Table 6.
Amino acid residues of Argonaute protein participating in the interaction with miR-106a, miR-21, and miR-29b-2 within a distance of 3.5 Å
| miRNA |
Amino acid residues |
||
|---|---|---|---|
| Hydrophobic interactionsa | With aromatic ringsb | Hydrogen bondingc | |
| miR-106a | LEU 132, ALA 133, VAL 152d, LEU 153d, ALA 170d, ILE 173d, LEU 267d, LEU 279e, ALA 479d, VAL 620d, VAL 663d, VAL 666, ILE 671 | TYR 135d, TYR 171d, TRP 415e, PHE 487 | SER 107, TYR 135, LEU 153, PRO 169, TYR 171, GLU 416, PHE 487, ARG 548d, ARG 574d, ASP 598d, ARG 615e, LYS 664d |
| miR-21 | ILE 173d, VAL 264, LEU 267d, LEU 279e, ALA 354, ALA 414d, ILE 434, ALA 644, ALA 648d, VAL 685 | TYR 135d, TYR 171d, TRP 415e, TYR 642d, PHE 647d, PHE 649d | ARG 172, ARG 192, THR 201, LEU 279, ARG 350, ALA 354, GLN 355, PRO 412d, MET 413, TRP 415, ARG 418, ASN 436, ARG 615e, ALA 644, PHE 649d, ARG 661d, ARG 668 |
| miR-29b-2 | VAL 152d, LEU 153d, ALA 170d, LEU 279e, ALA 414d, ALA 479d, LEU 596, VAL 620d, ALA 648d, LEU 652, ALA 659, LEU 662, VAL 663d | TRP 156, TRP 415e, TYR 642d, PHE 647d, PHE 649d | ARG 13, LYS 101, ARG 114, VAL 152, ASP 159, ARG 286, PRO 412d, ALA 414, ASP 478, ARG 548d, ARG 574d, LYS 575, |
| ARG 580, GLU 597, ASP 598d, ARG 611, ARG 615e, ALA 648, PHE 649d, ARG 651, HIS 657, ASP 660, ARG 661d, LYS 664d | |||
Table 7.
Amino acid residues of Argonaute protein participating in hydrogen bonding with miRNAs within a distance of 2.5 Amino acid residues of Argonaute protein participating in hydrogen bonding with miRNAs within a distance of 2.5 Å
Table 8.
Docking scores between miRNAs-mRNA duplex and AGO protein
| miRNA-mRNA and AGO (PDB ID:3F73, chain A) | Score | Area | ACE |
|---|---|---|---|
| miR-106a_PTEN-AGO | 19,722 | 3,519.90 | –408.36 |
| miR-21_TGFBR2-AGO | 20,036 | 4,132.60 | –863.02 |
| miR-29b-2_VEGFA-AGO | 24,310 | 3,927.40 | –70.34 |
Table 9.
Amino acid residues of Argonaute protein participating during interaction with miRNA and mRNA duplex within a distance of 3.5 Å
| miRNA-mRNA duplex |
Amino acid residues |
|
|---|---|---|
| Hydrophobic interactionsa | With aromatic ringsb | |
| miR-106a and PTEN | VAL 152c, LEU 153c, VAL 264c, LEU 265c, LEU 267c, LEU 277c, ALA 278c, LEU 279c, ALA 414, ILE 434, ALA 479c, VAL 663d, VAL 685c | TYR 43d, TRP 415d, PHE 487, TYR 642d |
| miR-21 and TGFBR2 | LEU 45, LEU 46, ALA 47, VAL 49, ALA 50, ALA 80c, ILE 173c, LEU 265c, LEU 267c, LEU 277c, ALA 278c, LEU 279c, LEU 281, ALA 479c, ALA 644c, ALA 648c, LEU 652, VAL 663d | TYR 43d, TRP 202, TRP 415d, TYR 642d, PHE 647c, PHE 649c |
| miR-29b-2 and VEGFA | ALA 80c, LEU 132, VAL 152c, LEU 153c, ALA 170, ILE 173, VAL 264c, ALA 479, VAL 606, ALA 644c, ALA 648c, VAL 663d, VAL 685c | TYR 43d, TYR 86, TYR 135, TRP 415d, TYR 171, TYR 642d, PHE 647c, PHE 649c |
- TOOLS
-
METRICS

-
- 8 Crossref
- 0 Scopus
- 8,342 View
- 102 Download
- Related articles in GNI





