 |
 |
- Search
| Genomics Inform > Volume 12(4); 2014 > Article |
Abstract
Platelets are derived from the fragments that are formed from the cytoplasm of bone marrow megakaryocytes-small irregularly shaped anuclear cells. Platelets respond to vascular damage, contracts blood vessels, and attaches to the damaged region, thereby stopping bleeding, together with the action of blood coagulation factors. Platelet activation is known to affect genes associated with vascular risk factors, as well as with arteriosclerosis and myocardial infarction. Here, we performed a genome-wide association study with 352,228 single-nucleotide polymorphisms typed in 8,842 subjects of the Korea Association Resource (KARE) project and replicated the results in 7,861 subjects from an independent population. We identified genetic associations between platelet count and common variants nearby chromosome 4p16.1 (p = 1.46 ├Ś 10-10, in the KIAA0232 gene), 6p21 (p = 1.36 ├Ś 10-7, in the BAK1 gene), and 12q24.12 (p = 1.11 ├Ś 10-15, in the SH2B3 gene). Our results illustrate the value of large-scale discovery and a focus for several novel research avenues.
Blood circulates through the body, affecting the tissues by delivering oxygen and nutrients responsible for tissue viability. The size, number, and concentration of a cell in the blood vary among populations, and these have been regarded as factors influencing various disorders, such as erythrocytosis, anemia, hypertension, and cardiovascular diseases. During the last 2 decades, genetic studies have revealed that there exists a genetic effect on several hematologic variables. Moreover, previous studies reported that genetic factors strongly influence the variation in the counts and size of blood cells [1, 2]. Although a few genes are known to affect hematologic traits, the physiological mechanisms underlying the traits are largely unrevealed.
Platelets are anuclear cytoplasmic fragments that play a key role in maintaining primary adhesion, aggregation, and secretion and providing procoagulant surface and clot retraction. The primary functions of a platelet count are to assist in the diagnosis of bleeding disorders and to monitor patients who are being treated for any disease involving bone marrow failure. Low platelet counts or abnormally shaped platelets are associated with bleeding disorders, whereas high platelet counts sometimes indicate disorders of the bone marrow. Platelet activation is known to affect genes associated with vascular risk factors, as well as with arteriosclerosis and myocardial infarction. Platelet count is a readily available laboratory test and has been associated with different clinical and epidemiologic factors and are tightly regulated and inversely correlated in the healthy population [3]. Furthermore, because platelet count differs between inter-ethnic groups, gender and ethnicity should be important considerations for a platelet count study [4].
Recently, genome-wide association studies (GWASs) for hematological traits have been reported [5, 6, 7]. Particularly, 68 single-nucleotide polymorphisms (SNPs) associated with platelet count were identified by a GWAS [8]. However, although the heritability of variation in platelet count ranged from 54% to more than 80% [9, 10, 11], the genetic variants reported to date explain only a small fraction of the heritability in platelet count [12]. Therefore, new studies have opportunities to unveil additional genetic variants for explaining missing heritability [12].
In this study, we conducted a GWAS to find a contribution of the genetic basis on hematologic variables, such as platelet count, in Koreans.
We conducted a GWAS using individuals sampled from a part of the Korean Genome Epidemiology Study (KoGES). The study subjects were recruited from population-based cohorts in the regions of Anseong and Ansan. The standardized examinations applied in this survey included 10,038 participants aged 40 to 69 years. Subjects for the replication study were recruited from the Health2 cohorts, another population-based cohort comprising the Wonju, Pyeongchang, Gangneung, Geumsan, and Naju regions in Korea. Among a total of 8,500 participants within the Health2 cohort, 7,861 subjects were selected for the replication analysis based on their age and information about concomitant disease and medication [13].
Peripheral blood was isolated by using the G-DEX TM IIb DNA Extraction Kit (iNtRON Biotechnology, Seongnam, Korea). Platelet count was measured using an ADVIA 120 (Bayer, Tarrytown, NY, USA). Genomic DNA, genotyped on the Affymetrix Genome-Wide Human SNP array 5.0 (Affymetrix, Inc., Santa Clara, CA, USA), was isolated from peripheral blood drawn from Anseong and Ansan cohort participants. Of 9,603 genotyped samples, we excluded samples with high heterozygosity (>30%, n = 11) and gender inconsistencies (n = 41). Also, individuals who had developed any kind of cancer (n = 101) were excluded from subsequent analyses. To examine population stratification overall, related or identical individuals with higher values than first-degree relatives of Korean sib-pair samples were also excluded according to average pair-wise identity-by-state (IBS) values (>0.80, n = 608). The methods to estimate heterozygosity and IBS have been described elsewhere [13]. SNP markers with a high missing genotype rate (>5%), low minor allele frequency (MAF, <0.01), and significant deviation from Hardy-Weinberg equilibrium (p < 1 ├Ś 10-6) were excluded. Consequently, a total of 352,228 markers were used for the GWAS. For the replication analysis, 19 SNPs of the Health2 population, comprising 7,861 participants, were genotyped by GoldenGate assay (Illumina Inc., San Diego, CA, USA).
Imputation was carried out by using the IMPUTE program version 1 (http://mathgen.stats.ox.ac.uk/impute/impute.html) [14, 15]. On the basis of NCBI, build 36 and dbSNP, build 126, we initially used 90 individuals from Japanese in Tokyo, Japan (JPT) and Han Chinese in Beijing, China (CHB) founders in HapMap as a reference panel, comprising 3.99 million SNPs (release 22). After removing SNPs with an MAF < 0.01 and an SNP missing rate > 0.05, we combined the remaining 1.8 million imputed SNPs with the directly typed Korea Association Resource (KARE) SNPs for the association analyses. Association analyses for imputed SNPs were carried out by the SNPTEST program (http://www.stats.ox.ac.uk/~marchini/software/gwas/snptest.html).
Association analysis of platelet trait with genotypes was performed using a linear regression model, adjusting for age, sex, and recruitment area. Analyses were performed with the software PLINK (http://pngu.mgh.harvard.edu/~purcell/plink), SAS version 9.1 (SAS Inc., Cary, NC, USA), and R statistics package version 2.7.1. The KARE GWAS and replication study were combined by an inverse-variance meta-analysis method, assuming fixed effects, with Cochran's Q test to assess between-study heterogeneity [16]. Regional plots were generated using LocusZoom [17].
Characteristics of the study participants, including age, sex, and trait summaries, are presented in Table 1. Quantile-quantile analysis of 2-d.f. logistic regression statistics for the comparison of genotype frequencies in the cohorts confirmed the genetic homogeneity of these two components of the KARE study population (Fig. 1). We observed significant SNPs by applying a Bonferroni adjustment cut-off with a combined p-value < 1.0 ├Ś 10-7. The GWAS identified several genomic locations as potentially associated with platelet count (Fig. 2). In a follow-up examination of the Health2 cohort, we examined 7,861 selected from 8,500 participants (aged, 40 to 69 years). We confirmed 3 replicated signals, of which 3 signals, replicating previously documented reports, and one novel locus were discovered.
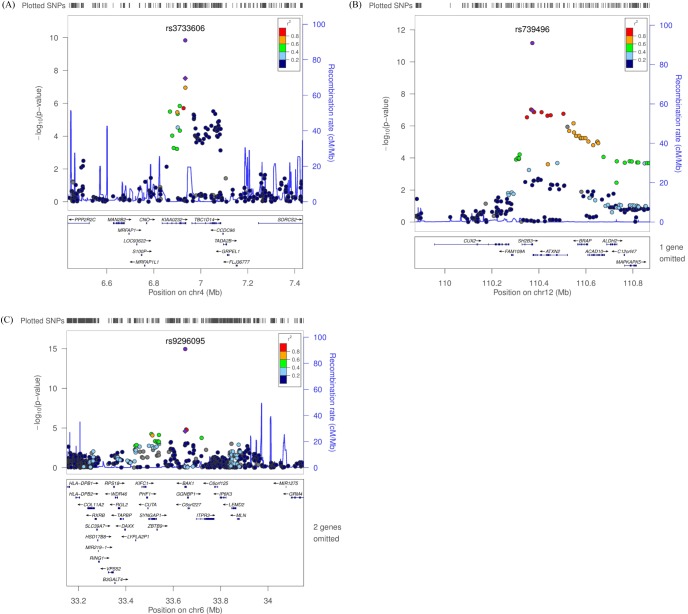
We observed 3 independent signals with p < 10-5 for platelet counts. Table 2 and Fig. 3 show results for three regions that had combined genomewide significant evidence for association with platelet count in the KARE GWAS and replication samples. For platelet counts, the strongest evidence for association was at 12q24 (rs739496, MAF = 0.11, combined p = 1.15 ├Ś 10-15) (Table 2, Fig. 3B). The associated signal is concordant with a previously published result in the Japanese population [6]. This SNP is located in the 3' untranslated region (UTR) of SH2B adaptor protein 3 (also known as LNK). SH2B3 is a member of the APS family of adaptor proteins, which play a pivotal role as broad inhibitors of growth factors and cytokine signaling pathways. The second SNP, rs3733606 (MAF = 0.50, combined p = 1.46 ├Ś 10-10) (Table 2, Fig. 3A) in 4p16, is in the 3' UTR of the KIAA0232 gene, which translates the functionally unknown hypothetical protein LOC9778. The third locus associated with platelet is at 6p21 (rs9296095, MAF = 0.23, combined p = 1.67 ├Ś 10-7) (Table 2, Fig. 3C).
We performed a GWAS of platelet count using 352,225 SNPs profiled with the Affymetrix Genome-Wide human SNP array 5.0 in 8,842 individuals from the Anseong and Ansan cohorts as described previously [13]. In a two-stage design (8,842 discovery and 7,861 replication samples), we confirmed three loci associated with platelet count at a genomewide significance level (< 1.0 ├Ś 10-7). Besides an unknown functional gene, we found two candidate genes, SH2B3 and BAK1, responsible for the variation in platelet counts. These genes are potential candidates for affecting platelet count.
Lymphocyte adapter protein (SH2B3, also known as LNK) is expressed in hematopoietic precursor cells and in endothelial cells and is known to be involved in inflammation [18]. LNK-deficient mice show a marked difference of hematopoiesis, accompanying increased numbers of various types of cells, such as megakaryocytes, B lymphoid, erythroid progenitor, and hematopoietic stem cells [19]. Several SNPs within the SH2B3 region are well known as variants associated with blood pressure, myocardial infarction, type 1 diabetes, and celiac disease [20].
BAK1, a multidomain pro-apoptotic family member, has shown to be involved in apoptotic cell death, playing a role as an essential mediator [21, 22, 23, 24]. Kamatani et al. [6] reported that BAK1 is a putative strong candidate gene accounting for numbers of platelets. This SNP is located in the 4th intron in BAK1 (Bcl2-antagonist/killer1), which encodes a protein acting as a strong proapoptotic effector that is known to control platelet lifespan [25]. The intrinsic machinery for apoptosis regulates the life span of anucleate platelets [25].
KIAA0232 has no known biological function and no clue for a related biological pathway. Given the weak linkage disequilibrium block around KIAA0232, further investigation is required for the biological function.
Some studies have discovered genetic factors associated with platelet count through genome-wide associated studies across diverse ethnic groups [12, 26]. Qayyum et al. [12] reported that a candidate gene, BAK1, was significant in African-Americans. Also, Shameer et al. [26] reported that SH2B3 and KIAA0232 in European ancestry were also significant in platelet count and mean platelet volume, respectively.
Numerous previous studies have demonstrated the association between platelet counts and various phenotypes in human and mice [27]. The Atherosclerosis Risk in Community (ARIC) study has shown that platelet counts are positively correlated with leukocytes [28]. Turakhia et al. [29] also reported the association between higher platelet counts and residual thrombus after fibrinolytic therapy, which is in agreement with the ARIC study. The evidence of a relationship between platelet count and insulin resistance in non-obese type 2 diabetic patients was reported from a study on Japanese [30]. The number of platelets is also a possible predictor of the risk of death and cardiovascular disease [31].
In conclusion, we identified and validated common variants at 1 novel locus, BAK1, and 2 known variants, SH2B and KIAA0232, responsible for the variation of platelet counts in population-based cohorts. Our research demonstrates the results from a meta-analysis and follow-up genotyping to retrieve positive evidence for the association of 3 loci with platelet counts. In addition, fine mapping and functional studies on the discovered loci will help us understand the hidden physiological mechanisms underlying platelet count.
Acknowledgments
This work was supported by grants from the Korea Centers for Disease Control and Prevention (4845-301) and an intramural grant from the Korea National Institute of Health (2012-N73002-00).
References
1. Whitfield JB, Martin NG. Genetic and environmental influences on the size and number of cells in the blood. Genet Epidemiol 1985;2:133ŌĆō144. PMID: 4054596.


2. Garner C, Tatu T, Reittie JE, Littlewood T, Darley J, Cervino S, et al. Genetic influences on F cells and other hematologic variables: a twin heritability study. Blood 2000;95:342ŌĆō346. PMID: 10607722.


3. Soranzo N, Rendon A, Gieger C, Jones CI, Watkins NA, Menzel S, et al. A novel variant on chromosome 7q22.3 associated with mean platelet volume, counts, and function. Blood 2009;113:3831ŌĆō3837. PMID: 19221038.



4. Bain BJ. Ethnic and sex differences in the total and differential white cell count and platelet count. J Clin Pathol 1996;49:664ŌĆō666. PMID: 8881919.



5. Chen Z, Tang H, Qayyum R, Schick UM, Nalls MA, Handsaker R, et al. Genome-wide association analysis of red blood cell traits in African Americans: the COGENT Network. Hum Mol Genet 2013;22:2529ŌĆō2538. PMID: 23446634.



6. Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet 2010;42:210ŌĆō215. PMID: 20139978.


7. Soranzo N, Spector TD, Mangino M, K├╝hnel B, Rendon A, Teumer A, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet 2009;41:1182ŌĆō1190. PMID: 19820697.



8. Chami N, Lettre G. Lessons and implications from genome-wide association studies (GWAS) findings of blood cell phenotypes. Genes (Basel) 2014;5:51ŌĆō64. PMID: 24705286.



9. Biino G, Balduini CL, Casula L, Cavallo P, Vaccargiu S, Parracciani D, et al. Analysis of 12,517 inhabitants of a Sardinian geographic isolate reveals that predispositions to thrombocytopenia and thrombocytosis are inherited traits. Haematologica 2011;96:96ŌĆō101. PMID: 20823129.



10. Buckley MF, James JW, Brown DE, Whyte GS, Dean MG, Chesterman CN, et al. A novel approach to the assessment of variations in the human platelet count. Thromb Haemost 2000;83:480ŌĆō484. PMID: 10744157.


11. Evans DM, Frazer IH, Martin NG. Genetic and environmental causes of variation in basal levels of blood cells. Twin Res 1999;2:250ŌĆō257. PMID: 10723803.


12. Qayyum R, Snively BM, Ziv E, Nalls MA, Liu Y, Tang W, et al. A meta-analysis and genome-wide association study of platelet count and mean platelet volume in african americans. PLoS Genet 2012;8:e1002491. PMID: 22423221.



13. Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet 2009;41:527ŌĆō534. PMID: 19396169.


14. Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661ŌĆō678. PMID: 17554300.




15. Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007;39:906ŌĆō913. PMID: 17572673.


16. Ioannidis JP, Patsopoulos NA, Evangelou E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One 2007;2:e841. PMID: 17786212.



17. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010;26:2336ŌĆō2337. PMID: 20634204.



18. Fitau J, Boulday G, Coulon F, Quillard T, Charreau B. The adaptor molecule Lnk negatively regulates tumor necrosis factor-alpha-dependent VCAM-1 expression in endothelial cells through inhibition of the ERK1 and -2 pathways. J Biol Chem 2006;281:20148ŌĆō20159. PMID: 16644735.


19. Velazquez L, Cheng AM, Fleming HE, Furlonger C, Vesely S, Bernstein A, et al. Cytokine signaling and hematopoietic homeostasis are disrupted in Lnk-deficient mice. J Exp Med 2002;195:1599ŌĆō1611. PMID: 12070287.



20. Ganesh SK, Zakai NA, van Rooij FJ, Soranzo N, Smith AV, Nalls MA, et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet 2009;41:1191ŌĆō1198. PMID: 19862010.



21. Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, et al. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell 2001;8:705ŌĆō711. PMID: 11583631.


22. Lindsten T, Ross AJ, King A, Zong WX, Rathmell JC, Shiels HA, et al. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol Cell 2000;6:1389ŌĆō1399. PMID: 11163212.



23. Rathmell JC, Lindsten T, Zong WX, Cinalli RM, Thompson CB. Deficiency in Bak and Bax perturbs thymic selection and lymphoid homeostasis. Nat Immunol 2002;3:932ŌĆō939. PMID: 12244308.


24. Zong WX, Lindsten T, Ross AJ, MacGregor GR, Thompson CB. BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev 2001;15:1481ŌĆō1486. PMID: 11410528.



25. Mason KD, Carpinelli MR, Fletcher JI, Collinge JE, Hilton AA, Ellis S, et al. Programmed anuclear cell death delimits platelet life span. Cell 2007;128:1173ŌĆō1186. PMID: 17382885.


26. Shameer K, Denny JC, Ding K, Jouni H, Crosslin DR, de Andrade M, et al. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum Genet 2014;133:95ŌĆō109. PMID: 24026423.



27. Cheung CC, Martin IC, Zenger KR, Donald JA, Thomson PC, Moran C, et al. Quantitative trait loci for steady-state platelet count in mice. Mamm Genome 2004;15:784ŌĆō797. PMID: 15520881.


28. Nieto FJ, Szklo M, Folsom AR, Rock R, Mercuri M. Leukocyte count correlates in middle-aged adults: the Atherosclerosis Risk in Communities (ARIC) Study. Am J Epidemiol 1992;136:525ŌĆō537. PMID: 1442716.


29. Turakhia MP, Murphy SA, Pinto TL, Antman EM, Giugliano RP, Cannon CP, et al. Association of platelet count with residual thrombus in the myocardial infarct-related coronary artery among patients treated with fibrinolytic therapy for ST-segment elevation acute myocardial infarction. Am J Cardiol 2004;94:1406ŌĆō1410. PMID: 15566912.


30. Taniguchi A, Fukushima M, Seino Y, Sakai M, Yoshii S, Nagasaka S, et al. Platelet count is independently associated with insulin resistance in non-obese Japanese type 2 diabetic patients. Metabolism 2003;52:1246ŌĆō1249. PMID: 14564674.


31. Thaulow E, Erikssen J, Sandvik L, Stormorken H, Cohn PF. Blood platelet count and function are related to total and cardiovascular death in apparently healthy men. Circulation 1991;84:613ŌĆō617. PMID: 1860204.


Fig.┬Ā1
Quantile-quantile plot for platelet count. The observed p-values (y axis) were compared with the expected p-values under the null distribution (x axis) for each trait. The shaded region represents the 95% concentration band.
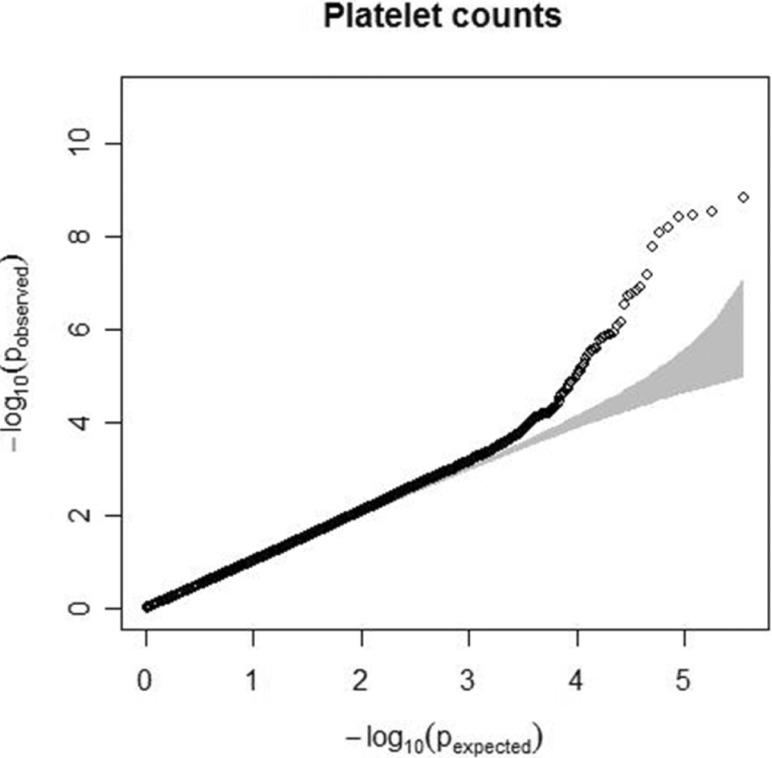
Fig.┬Ā2
Chromosome plot for platelet counts. Genome-wide association study for log-transformed platelet count on a population-based sample of 8,842 individuals from the Korea Association Resource (KARE) study. The x axis represents the genomic position (in Gb) of 352,225 single nucleotide polymorphisms; they show -log10(p-value). Single nucleotide polymorphisms with a p-value < 1 ├Ś 10-4 are highlighted in red.

Fig.┬Ā3
Regional plot of three discovered variants. (A-C) p-value plots showing the association signals in the region of KIAA0232 on chromosome 4 (A), SH2B3 on chromosome 12 (B), and BAK1 on chromosome 6 (C). In the top panel, the association signals scaled by -log10(p-value) (typed or imputed SNPs) at each locus are distributed in a genomic region 500 kb to either side of the lead association signal (typed). Each SNP is plotted as a circle along the chromosomal position, and linkage disequilibrium between the lead SNP and the other SNPs is colored as a scale from low (blue) to high (red) or is colored gray if linkage disequilibrium information was not available in the 1,000 genomes June 2010 CHB+JPT samples. The lead SNP is colored purple diamond, and the overall meta-analysis result is shown with a purple circle. The recombination rate estimated from HapMap phase 2 is plotted in blue. The bottom panel illustrates the locations of known genes. Genetic information is based on NCBI build 36 and dbSNP build 130. SNP, single-nucleotide polymorphism; CHB, Han Chinese in Beijing, China; JPT, Japanese in Tokyo, Japan.
Table┬Ā1.
Samples used in this study
Table┬Ā2.
Variants that associate with variation in platelet counts
- TOOLS
-
METRICS

-
- 13 Crossref
- 6,461 View
- 71 Download
- Related articles in GNI





