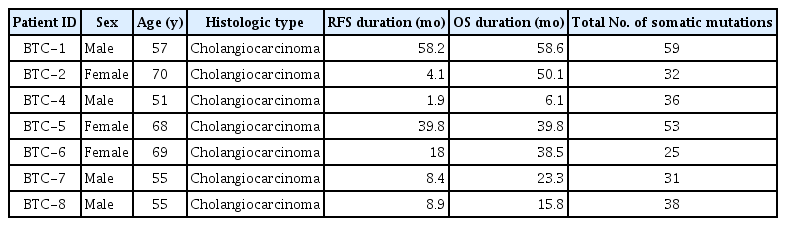
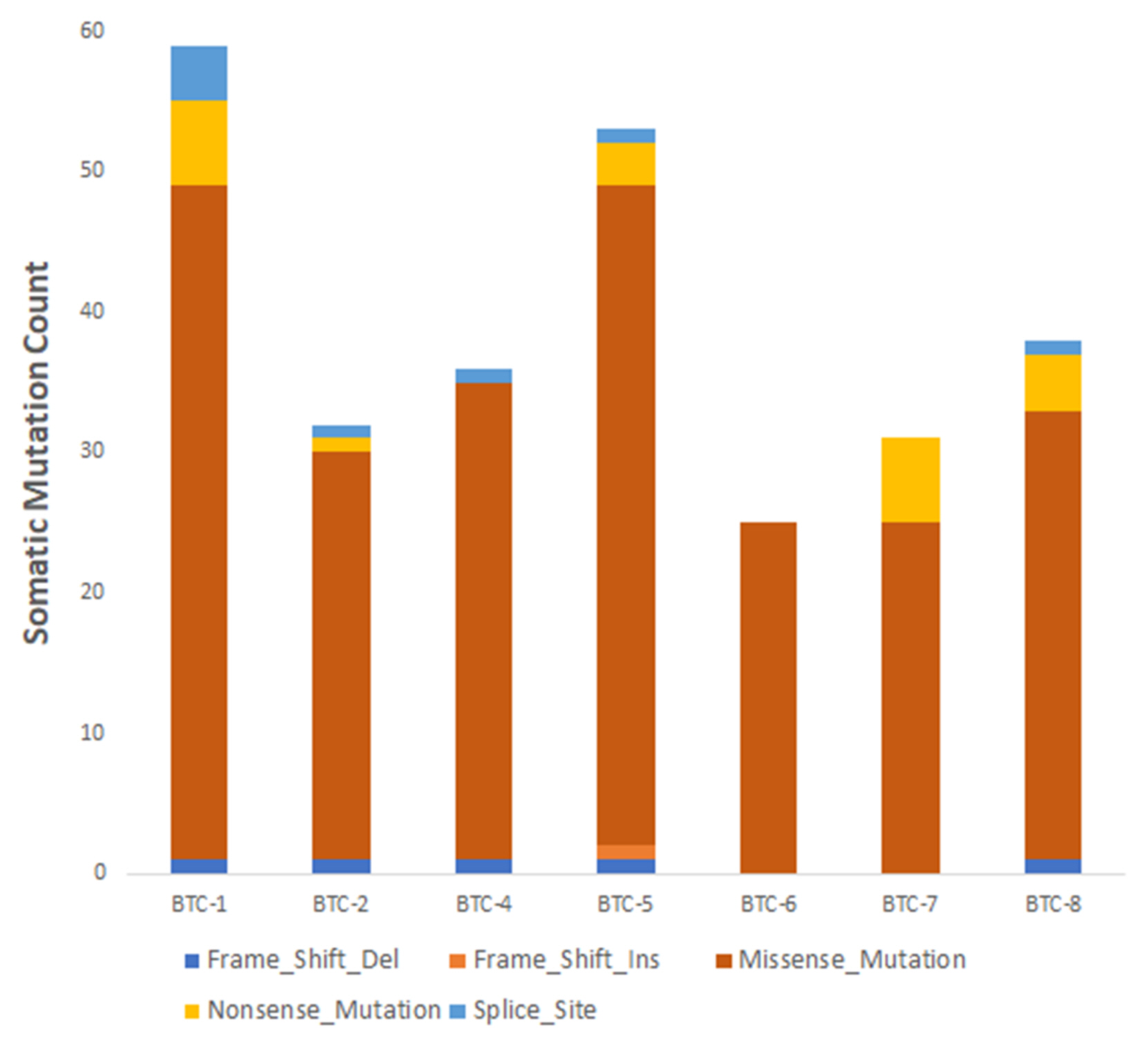
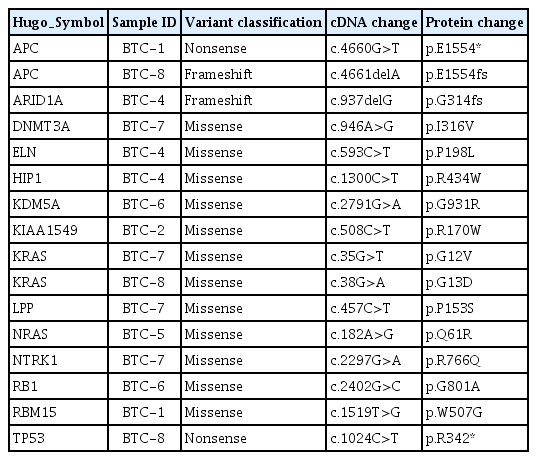
| BTC-1 |
APC |
324 |
5 |
112174759 |
112174760 |
Frame_Shift_Del |
AG |
- |
c.3468_3469delAG |
c.(3466-34p.EE1156fs |
adenomatous polyposis coli |
| BTC-1 |
KRTAP19-6 |
337973 |
21 |
31914024 |
31914024 |
Nonsense_Mutation |
G |
T |
c.129C>A |
c.(127-129)p.C43* |
keratin associated protein 19-6 |
| BTC-1 |
PABPC4L |
132430 |
4 |
135121319 |
135121319 |
Nonsense_Mutation |
G |
A |
c.856C>T |
c.(856-858)p.R286* |
poly(A) binding protein, cytoplasmic 4-like |
| BTC-1 |
APC |
324 |
5 |
112175951 |
112175951 |
Nonsense_Mutation |
G |
T |
c.4660G>T |
c.(4660-46p.E1554* |
adenomatous polyposis coli |
| BTC-1 |
C6orf222 |
389384 |
6 |
36290159 |
36290159 |
Nonsense_Mutation |
G |
C |
c.1532C>G |
c.(1531-15p.S511* |
chromosome 6 open reading frame 222 |
| BTC-1 |
ABCA1 |
19 |
9 |
107553269 |
107553269 |
Nonsense_Mutation |
G |
C |
c.5861C>G |
c.(5860-58p.S1954* |
ATP-binding cassette, sub-family A (ABC1), member 1 |
| BTC-1 |
AMER1 |
139285X |
|
63412530 |
63412530 |
Nonsense_Mutation |
G |
A |
c.637C>T |
c.(637-639)p.Q213* |
APC membrane recruitment protein 1 |
| BTC-1 |
MFAP2 |
4237 |
1 |
17302995 |
17302995 |
Splice_Site |
C |
A |
|
c.e5+1 |
microfibrillar-associated protein 2 |
| BTC-1 |
HIF3A |
64344 |
19 |
46825222 |
46825222 |
Splice_Site |
C |
T |
c.1334C>T |
c.(1333-13p.S445L |
hypoxia inducible factor 3, alpha subunit |
| BTC-1 |
MAP3K13 |
9175 |
3 |
185155234 |
185155234 |
Splice_Site |
G |
C |
|
c.e3-1 |
mitogen-activated protein kinase kinase kinase 13 |
| BTC-1 |
DAB2IP |
153090 |
9 |
124329507 |
124329507 |
Splice_Site |
G |
A |
c.40G>A |
c.(40-42)Gap.E14K |
DAB2 interacting protein |
| BTC-1 |
ABCC2 |
1244 |
10 |
101591752 |
101591752 |
Missense_Mutation |
C |
T |
c.3122C>T |
c.(3121-31p.A1041V |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| BTC-1 |
ACE |
1636 |
17 |
61570945 |
61570945 |
Missense_Mutation |
G |
A |
c.1339G>A |
c.(1339-134p.V447M |
angiotensin I converting enzyme |
| BTC-1 |
ACTN2 |
88 |
1 |
236899007 |
236899007 |
Missense_Mutation |
C |
T |
c.770C>T |
c.(769-771)p.A257V |
actinin, alpha 2 |
| BTC-1 |
ACTN4 |
81 |
19 |
39207832 |
39207832 |
Missense_Mutation |
G |
A |
c.1019G>A |
c.(1018-10p.R340H |
actinin, alpha 4 |
| BTC-1 |
ACVR1B |
91 |
12 |
52379016 |
52379016 |
Missense_Mutation |
C |
A |
c.864C>A |
c.(862-864)p.N288K |
activin A receptor, type IB |
| BTC-1 |
ADCY5 |
111 |
3 |
123166420 |
123166420 |
Missense_Mutation |
C |
T |
c.973G>A |
c.(973-975)p.A325T |
adenylate cyclase 5 |
| BTC-1 |
ADCY8 |
114 |
8 |
131949376 |
131949376 |
Missense_Mutation |
C |
T |
c.1424G>A |
c.(1423-14p.R475H |
adenylate cyclase 8 (brain) |
| BTC-1 |
ANO3 |
63982 |
11 |
26463533 |
26463533 |
Missense_Mutation |
T |
C |
c.115T>C |
c.(115-117)p.C39R |
anoctamin 3 |
| BTC-1 |
APC2 |
10297 |
19 |
1457029 |
1457029 |
Missense_Mutation |
G |
A |
c.994G>A |
c.(994-996)p.A332T |
adenomatosis polyposis coli 2 |
| BTC-1 |
AQPEP |
0 |
5 |
115298621 |
115298621 |
Missense_Mutation |
G |
A |
c.307G>A |
c.(307-309)p.V103M |
|
| BTC-1 |
AUTS2 |
26053 |
7 |
69364311 |
69364311 |
Missense_Mutation |
C |
T |
c.349C>T |
c.(349-351)p.R117C |
autism susceptibility candidate 2 |
| BTC-1 |
C11orf63 |
79864 |
11 |
122775018 |
122775018 |
Missense_Mutation |
C |
T |
c.730C>T |
c.(730-732)p.R244C |
chromosome 11 open reading frame 63 |
| BTC-1 |
C5orf42 |
65250 |
5 |
37153858 |
37153858 |
Missense_Mutation |
T |
A |
c.8195A>T |
c.(8194-81p.D2732V |
chromosome 5 open reading frame 42 |
| BTC-1 |
CDH2 |
1000 |
18 |
25572711 |
25572711 |
Missense_Mutation |
C |
T |
c.1159G>A |
c.(1159-11p.A387T |
cadherin 2, type 1, N-cadherin (neuronal) |
| BTC-1 |
CHST9 |
83539 |
18 |
24628462 |
24628462 |
Missense_Mutation |
C |
T |
c.127G>A |
c.(127-129)p.V43M |
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| BTC-1 |
CYP11B1 |
1584 |
8 |
143956558 |
143956558 |
Missense_Mutation |
C |
T |
c.1426G>A |
c.(1426-14p.V476M |
cytochrome P450, family 11, subfamily B, polypeptide 1 |
| BTC-1 |
DNAH10 |
196385 |
12 |
124341718 |
124341718 |
Missense_Mutation |
G |
A |
c.6200G>A |
c.(6199-62p.R2067H |
dynein, axonemal, heavy chain 10 |
| BTC-1 |
DSCAM |
1826 |
21 |
41559851 |
41559851 |
Missense_Mutation |
C |
T |
c.2617G>A |
c.(2617-26p.E873K |
Down syndrome cell adhesion molecule |
| BTC-1 |
ELF3 |
1999 |
1 |
201983140 |
201983140 |
Missense_Mutation |
G |
A |
c.989G>A |
c.(988-990)p.S330N |
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| BTC-1 |
FGD3 |
89846 |
9 |
95738639 |
95738639 |
Missense_Mutation |
C |
T |
c.101C>T |
c.(100-102)p.A34V |
FYVE, RhoGEF and PH domain containing 3 |
| BTC-1 |
FUBP3 |
8939 |
9 |
133493210 |
133493210 |
Missense_Mutation |
G |
A |
c.594G>A |
c.(592-594)p.M198I |
far upstream element (FUSE) binding protein 3 |
| BTC-1 |
HAND1 |
9421 |
5 |
153855452 |
153855452 |
Missense_Mutation |
G |
T |
c.562C>A |
c.(562-564)p.P188T |
heart and neural crest derivatives expressed 1 |
| BTC-1 |
KCNS2 |
3788 |
8 |
99440299 |
99440299 |
Missense_Mutation |
G |
A |
c.92G>A |
c.(91-93)cGp.R31H |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
| BTC-1 |
KIAA1429 |
25962 |
8 |
95503865 |
95503865 |
Missense_Mutation |
C |
G |
c.5081G>C |
c.(5080-50p.R1694T |
KIAA1429 |
| BTC-1 |
KRTAP19-1 |
337882 |
21 |
31852374 |
31852374 |
Missense_Mutation |
C |
T |
c.263G>A |
c.(262-264)p.G88D |
keratin associated protein 19-1 |
| BTC-1 |
LRP1B |
53353 |
2 |
141004693 |
141004693 |
Missense_Mutation |
G |
T |
c.13286C>A |
c.(13285-1p.P4429Q |
low density lipoprotein receptor-related protein 1B |
| BTC-1 |
LRRK2 |
120892 |
12 |
40689336 |
40689336 |
Missense_Mutation |
G |
T |
c.2986G>T |
c.(2986-29p.D996Y |
leucine-rich repeat kinase 2 |
| BTC-1 |
MAP2K1 |
5604 |
15 |
66727455 |
66727455 |
Missense_Mutation |
G |
C |
c.171G>C |
c.(169-171)p.K57N |
mitogen-activated protein kinase kinase 1 |
| BTC-1 |
MBTD1 |
54799 |
17 |
49296318 |
49296318 |
Missense_Mutation |
A |
C |
c.376T>G |
c.(376-378)p.L126V |
mbt domain containing 1 |
| BTC-1 |
MUC19 |
283463 |
12 |
40820354 |
40820354 |
Missense_Mutation |
G |
A |
c.332G>A |
c.(331-333)p.R111H |
mucin 19, oligomeric |
| BTC-1 |
MYLK2 |
85366 |
20 |
30419645 |
30419645 |
Missense_Mutation |
G |
A |
c.1564G>A |
c.(1564-15p.V522I |
myosin light chain kinase 2 |
| BTC-1 |
PCDH17 |
27253 |
13 |
58207860 |
58207860 |
Missense_Mutation |
G |
A |
c.1180G>A |
c.(1180-11p.G394R |
protocadherin 17 |
| BTC-1 |
PLIN4 |
729359 |
19 |
4510530 |
4510530 |
Missense_Mutation |
G |
A |
c.3400C>T |
c.(3400-34p.R1134C |
perilipin 4 |
| BTC-1 |
PRRC2B |
84726 |
9 |
134322013 |
134322013 |
Missense_Mutation |
A |
G |
c.839A>G |
c.(838-840)p.D280G |
proline-rich coiled-coil 2B |
| BTC-1 |
RBM15 |
64783 |
1 |
110883546 |
110883546 |
Missense_Mutation |
T |
G |
c.1519T>G |
c.(1519-15p.W507G |
RNA binding motif protein 15 |
| BTC-1 |
RP1-139D8 |
0 |
6 |
42123292 |
42123292 |
Missense_Mutation |
G |
T |
c.14G>T |
c.(13-15)aGp.S5I |
|
| BTC-1 |
SDK1 |
221935 |
7 |
4273010 |
4273010 |
Missense_Mutation |
C |
T |
c.5891C>T |
c.(5890-58p.A1964V |
sidekick cell adhesion molecule 1 |
| BTC-1 |
SPRY2 |
10253 |
13 |
80911584 |
80911584 |
Missense_Mutation |
G |
C |
c.257C>G |
c.(256-258)p.P86R |
sprouty homolog 2 (Drosophila) |
| BTC-1 |
ST6GALNA |
81849 |
1 |
77509964 |
77509964 |
Missense_Mutation |
C |
A |
c.337C>A |
c.(337-339)p.Q113K |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| BTC-1 |
SYT3 |
84258 |
19 |
51135883 |
51135883 |
Missense_Mutation |
C |
T |
c.334G>A |
c.(334-336)p.G112R |
synaptotagmin III |
| BTC-1 |
TADA2A |
6871 |
17 |
35837062 |
35837062 |
Missense_Mutation |
G |
C |
c.1307G>C |
c.(1306-13p.R436T |
transcriptional adaptor 2A |
| BTC-1 |
TGM2 |
7052 |
20 |
36793594 |
36793594 |
Missense_Mutation |
C |
T |
c.7G>A |
c.(7-9)Gagp.E3K |
transglutaminase 2 |
| BTC-1 |
TMEM184B |
25829 |
22 |
38643871 |
38643871 |
Missense_Mutation |
T |
C |
c.97A>G |
c.(97-99)Acp.T33A |
transmembrane protein 184B |
| BTC-1 |
TRPC1 |
7220 |
3 |
142499703 |
142499703 |
Missense_Mutation |
A |
T |
c.792A>T |
c.(790-792)p.Q264H |
transient receptor potential cation channel, subfamily C, member 1 |
| BTC-1 |
TSHZ3 |
57616 |
19 |
31770589 |
31770589 |
Missense_Mutation |
G |
C |
c.110C>G |
c.(109-111)p.A37G |
teashirt zinc finger homeobox 3 |
| BTC-1 |
TSPAN14 |
81619 |
10 |
82271948 |
82271948 |
Missense_Mutation |
T |
G |
c.499T>G |
c.(499-501)p.Y167D |
tetraspanin 14 |
| BTC-1 |
ZFR |
51663 |
5 |
32395347 |
32395347 |
Missense_Mutation |
C |
T |
c.1897G>A |
c.(1897-18p.E633K |
zinc finger RNA binding protein |
| BTC-1 |
ZFYVE26 |
23503 |
14 |
68264958 |
68264958 |
Missense_Mutation |
C |
A |
c.2021G>T |
c.(2020-20p.G674V |
zinc finger, FYVE domain containing 26 |
| BTC-2 |
EPHA2 |
1969 |
1 |
16464447 |
16464447 |
Frame_Shift_Del |
G |
- |
c.1213delC |
c.(1213-12p.H405fs |
EPH receptor A2 |
| BTC-2 |
LGI4 |
163175 |
19 |
35617610 |
35617610 |
Nonsense_Mutation |
C |
T |
c.863G>A |
c.(862-864)p.W288* |
leucine-rich repeat LGI family, member 4 |
| BTC-2 |
EPHA2 |
1969 |
1 |
16456720 |
16456720 |
Splice_Site |
C |
T |
|
c.e15+1 |
EPH receptor A2 |
| BTC-2 |
C17orf97 |
400566 |
17 |
260315 |
260315 |
Missense_Mutation |
G |
T |
c.182G>T |
c.(181-183)p.G61V |
chromosome 17 open reading frame 97 |
| BTC-2 |
CAMK2N1 |
55450 |
1 |
20810156 |
20810156 |
Missense_Mutation |
G |
A |
c.223C>T |
c.(223-225)p.P75S |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| BTC-2 |
CNBD1 |
168975 |
8 |
88249206 |
88249206 |
Missense_Mutation |
G |
A |
c.637G>A |
c.(637-639)p.V213M |
cyclic nucleotide binding domain containing 1 |
| BTC-2 |
COL14A1 |
7373 |
8 |
121292323 |
121292323 |
Missense_Mutation |
G |
A |
c.3631G>A |
c.(3631-36p.E1211K |
collagen, type XIV, alpha 1 |
| BTC-2 |
FBN3 |
84467 |
19 |
8201283 |
8201283 |
Missense_Mutation |
C |
T |
c.1334G>A |
c.(1333-13p.G445D |
fibrillin 3 |
| BTC-2 |
FIGN |
55137 |
2 |
164467285 |
164467285 |
Missense_Mutation |
C |
A |
c.1057G>T |
c.(1057-10p.D353Y |
fidgetin |
| BTC-2 |
FLNB |
2317 |
3 |
58118580 |
58118580 |
Missense_Mutation |
A |
G |
c.4529A>G |
c.(4528-45p.H1510R |
filamin B, beta |
| BTC-2 |
INO80D |
54891 |
2 |
206884534 |
206884534 |
Missense_Mutation |
G |
A |
c.1334C>T |
c.(1333-13p.A445V |
INO80 complex subunit D |
| BTC-2 |
INTS1 |
26173 |
7 |
1544069 |
1544069 |
Missense_Mutation |
C |
G |
c.233G>C |
c.(232-234)p.C78S |
integrator complex subunit 1 |
| BTC-2 |
KCNIP1 |
30820 |
5 |
170145809 |
170145809 |
Missense_Mutation |
G |
A |
c.109G>A |
c.(109-111)p.E37K |
Kv channel interacting protein 1 |
| BTC-2 |
KIAA1549 |
57670 |
7 |
138603864 |
138603864 |
Missense_Mutation |
G |
A |
c.508C>T |
c.(508-510)p.R170W |
KIAA1549 |
| BTC-2 |
LGALS9 |
3965 |
17 |
25975970 |
25975970 |
Missense_Mutation |
G |
A |
c.934G>A |
c.(934-936)p.V312M |
lectin, galactoside-binding, soluble, 9 |
| BTC-2 |
PLEKHG1 |
57480 |
6 |
151152904 |
151152904 |
Missense_Mutation |
G |
A |
c.2657G>A |
c.(2656-26p.R886H |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| BTC-2 |
PNCK |
139728X |
|
152937421 |
152937421 |
Missense_Mutation |
C |
G |
c.328G>C |
c.(328-330)p.E110Q |
pregnancy up-regulated nonubiquitous CaM kinase |
| BTC-2 |
PPDPF |
79144 |
20 |
62152924 |
62152924 |
Missense_Mutation |
G |
A |
c.115G>A |
c.(115-117)p.A39T |
pancreatic progenitor cell differentiation and proliferation factor |
| BTC-2 |
PROKR2 |
128674 |
20 |
5282783 |
5282783 |
Missense_Mutation |
C |
T |
c.1058G>A |
c.(1057-10p.R353H |
prokineticin receptor 2 |
| BTC-2 |
RP11-429E |
0 |
20 |
60294088 |
60294088 |
Missense_Mutation |
G |
A |
c.139C>T |
c.(139-141)p.L47F |
|
| BTC-2 |
SCN5A |
6331 |
3 |
38645514 |
38645514 |
Missense_Mutation |
C |
T |
c.1579G>A |
c.(1579-15p.G527R |
sodium channel, voltage-gated, type V, alpha subunit |
| BTC-2 |
SLC35F1 |
222553 |
6 |
118556702 |
118556702 |
Missense_Mutation |
G |
A |
c.380G>A |
c.(379-381)p.R127Q |
solute carrier family 35, member F1 |
| BTC-2 |
SLC9A6 |
10479X |
|
135067950 |
135067950 |
Missense_Mutation |
C |
T |
c.289C>T |
c.(289-291)p.R97C |
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6 |
| BTC-2 |
SPARC |
6678 |
5 |
151047056 |
151047056 |
Missense_Mutation |
T |
C |
c.557A>G |
c.(556-558)p.N186S |
secreted protein, acidic, cysteine-rich (osteonectin) |
| BTC-2 |
TLL2 |
7093 |
10 |
98144434 |
98144434 |
Missense_Mutation |
C |
G |
c.2104G>C |
c.(2104-21p.V702L |
tolloid-like 2 |
| BTC-2 |
TRIM58 |
25893 |
1 |
248028097 |
248028097 |
Missense_Mutation |
C |
T |
c.607C>T |
c.(607-609)p.R203W |
tripartite motif containing 58 |
| BTC-2 |
TSPAN16 |
26526 |
19 |
11408947 |
11408947 |
Missense_Mutation |
A |
G |
c.199A>G |
c.(199-201)p.I67V |
tetraspanin 16 |
| BTC-2 |
TSSK2 |
23617 |
22 |
19119034 |
19119034 |
Missense_Mutation |
A |
C |
c.122A>C |
c.(121-123)p.K41T |
testis-specific serine kinase 2 |
| BTC-2 |
VAV2 |
7410 |
9 |
136635595 |
136635595 |
Missense_Mutation |
G |
A |
c.2222C>T |
c.(2221-22p.S741L |
vav 2 guanine nucleotide exchange factor |
| BTC-2 |
ZC2HC1B |
153918 |
6 |
144224199 |
144224199 |
Missense_Mutation |
C |
T |
c.508C>T |
c.(508-510)p.P170S |
zinc finger, C2HC-type containing 1B |
| BTC-2 |
ZFP69B |
65243 |
1 |
40928270 |
40928270 |
Missense_Mutation |
G |
A |
c.614G>A |
c.(613-615)p.G205D |
ZFP69 zinc finger protein B |
| BTC-2 |
ZNF423 |
23090 |
16 |
49669726 |
49669726 |
Missense_Mutation |
C |
T |
c.3337G>A |
c.(3337-33p.A1113T |
zinc finger protein 423 |
| BTC-4 |
ARID1A |
8289 |
1 |
27023831 |
27023831 |
Frame_Shift_Del |
G |
- |
c.937delG |
c.(937-939)p.G314fs |
AT rich interactive domain 1A (SWI-like) |
| BTC-4 |
TULP1 |
7287 |
6 |
35480415 |
35480415 |
Splice_Site |
C |
G |
|
c.e2+1 |
tubby like protein 1 |
| BTC-4 |
ACOXL |
55289 |
2 |
111850501 |
111850501 |
Missense_Mutation |
T |
A |
c.1590T>A |
c.(1588-15p.H530Q |
acyl-CoA oxidase-like |
| BTC-4 |
CACNA1H |
8912 |
16 |
1270626 |
1270626 |
Missense_Mutation |
C |
A |
c.6676C>A |
c.(6676-66p.L2226M |
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| BTC-4 |
CATSPERB |
79820 |
14 |
92174496 |
92174496 |
Missense_Mutation |
T |
A |
c.455A>T |
c.(454-456)p.D152V |
catsper channel auxiliary subunit beta |
| BTC-4 |
CDKN2A |
1029 |
9 |
21971035 |
21971035 |
Missense_Mutation |
T |
C |
c.170A>G |
c.(169-171)p.D57G |
cyclin-dependent kinase inhibitor 2A |
| BTC-4 |
CES3 |
23491 |
16 |
67006620 |
67006620 |
Missense_Mutation |
G |
A |
c.408G>A |
c.(406-408)p.M136I |
carboxylesterase 3 |
| BTC-4 |
CLCNKA |
1187 |
1 |
16349137 |
16349137 |
Missense_Mutation |
G |
A |
c.23G>A |
c.(22-24)cGp.R8H |
chloride channel, voltage-sensitive Ka |
| BTC-4 |
COL22A1 |
169044 |
8 |
139890232 |
139890232 |
Missense_Mutation |
C |
T |
c.419G>A |
c.(418-420)p.R140H |
collagen, type XXII, alpha 1 |
| BTC-4 |
CRISPLD1 |
83690 |
8 |
75941665 |
75941665 |
Missense_Mutation |
G |
A |
c.1364G>A |
c.(1363-13p.R455Q |
cysteine-rich secretory protein LCCL domain containing 1 |
| BTC-4 |
DAW1 |
164781 |
2 |
228767727 |
228767727 |
Missense_Mutation |
T |
C |
c.550T>C |
c.(550-552)p.S184P |
dynein assembly factor with WDR repeat domains 1 |
| BTC-4 |
DYSF |
8291 |
2 |
71797023 |
71797023 |
Missense_Mutation |
G |
A |
c.2938G>A |
c.(2938-294p.G980R |
dysferlin |
| BTC-4 |
ELN |
2006 |
7 |
73461047 |
73461047 |
Missense_Mutation |
C |
T |
c.593C>T |
c.(592-594)p.P198L |
elastin |
| BTC-4 |
FAM135B |
51059 |
8 |
139165339 |
139165339 |
Missense_Mutation |
A |
G |
c.1379T>C |
c.(1378-13p.L460P |
family with sequence similarity 135, member B |
| BTC-4 |
FAM208B |
54906 |
10 |
5803432 |
5803432 |
Missense_Mutation |
C |
A |
c.7172C>A |
c.(7171-71p.T2391K |
family with sequence similarity 208, member B |
| BTC-4 |
GPR65 |
8477 |
14 |
88478153 |
88478153 |
Missense_Mutation |
G |
A |
c.962G>A |
c.(961-963)p.R321H |
G protein-coupled receptor 65 |
| BTC-4 |
GUCY1A3 |
2982 |
4 |
156643206 |
156643206 |
Missense_Mutation |
A |
T |
c.1733A>T |
c.(1732-17p.H578L |
guanylate cyclase 1, soluble, alpha 3 |
| BTC-4 |
HID1 |
283987 |
17 |
72959068 |
72959068 |
Missense_Mutation |
T |
A |
c.496A>T |
c.(496-498)p.S166C |
HID1 domain containing |
| BTC-4 |
HIP1 |
3092 |
7 |
75189111 |
75189111 |
Missense_Mutation |
G |
A |
c.1300C>T |
c.(1300-13p.R434W |
huntingtin interacting protein 1 |
| BTC-4 |
IRF3 |
3661 |
19 |
50167939 |
50167939 |
Missense_Mutation |
T |
C |
c.157A>G |
c.(157-159)p.I53V |
interferon regulatory factor 3 |
| BTC-4 |
KCNB1 |
3745 |
20 |
48098906 |
48098906 |
Missense_Mutation |
C |
T |
c.112G>A |
c.(112-114)p.G38R |
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| BTC-4 |
KCNT1 |
57582 |
9 |
138664603 |
138664603 |
Missense_Mutation |
C |
T |
c.2051C>T |
c.(2050-20p.T684M |
potassium channel, subfamily T, member 1 |
| BTC-4 |
LLGL1 |
3996 |
17 |
18143990 |
18143990 |
Missense_Mutation |
C |
T |
c.2305C>T |
c.(2305-23p.R769W |
lethal giant larvae homolog 1 (Drosophila) |
| BTC-4 |
LPHN1 |
22859 |
19 |
14294398 |
14294398 |
Missense_Mutation |
G |
A |
c.17C>T |
c.(16-18)gCp.A6V |
latrophilin 1 |
| BTC-4 |
MARS |
4141 |
12 |
57906626 |
57906626 |
Missense_Mutation |
A |
T |
c.1846A>T |
c.(1846-184p.I616F |
methionyl-tRNA synthetase |
| BTC-4 |
MBTPS1 |
8720 |
16 |
84093019 |
84093019 |
Missense_Mutation |
G |
A |
c.2719C>T |
c.(2719-27p.R907W |
membrane-bound transcription factor peptidase, site 1 |
| BTC-4 |
PDC |
5132 |
1 |
186413572 |
186413572 |
Missense_Mutation |
G |
A |
c.280C>T |
c.(280-282)p.R94C |
phosducin |
| BTC-4 |
PTTG1IP |
754 |
21 |
46285352 |
46285352 |
Missense_Mutation |
C |
G |
c.126G>C |
c.(124-126)p.Q42H |
pituitary tumor-transforming 1 interacting protein |
| BTC-4 |
RBBP6 |
5930 |
16 |
24581304 |
24581304 |
Missense_Mutation |
A |
T |
c.3293A>T |
c.(3292-32p.E1098V |
retinoblastoma binding protein 6 |
| BTC-4 |
RBBP7 |
5931X |
|
16871823 |
16871823 |
Missense_Mutation |
T |
A |
c.872A>T |
c.(871-873)p.D291V |
retinoblastoma binding protein 7 |
| BTC-4 |
RDH10 |
157506 |
8 |
74234976 |
74234976 |
Missense_Mutation |
A |
G |
c.833A>G |
c.(832-834)p.K278R |
retinol dehydrogenase 10 (all-trans) |
| BTC-4 |
RFX8 |
731220 |
2 |
102018971 |
102018971 |
Missense_Mutation |
C |
T |
c.1172G>A |
c.(1171-11p.R391Q |
RFX family member 8, lacking RFX DNA binding domain |
| BTC-4 |
SEMA6D |
80031 |
15 |
48058143 |
48058143 |
Missense_Mutation |
C |
T |
c.1505C>T |
c.(1504-15p.A502V |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| BTC-4 |
SLC6A17 |
388662 |
1 |
110734682 |
110734682 |
Missense_Mutation |
C |
A |
c.953C>A |
c.(952-954)p.A318D |
solute carrier family 6 (neutral amino acid transporter), member 17 |
| BTC-4 |
TMEM205 |
374882 |
19 |
11455971 |
11455971 |
Missense_Mutation |
G |
A |
c.221C>T |
c.(220-222)p.S74L |
transmembrane protein 205 |
| BTC-4 |
TRIM4 |
89122 |
7 |
99506284 |
99506284 |
Missense_Mutation |
T |
C |
c.719A>G |
c.(718-720)p.N240S |
tripartite motif containing 4 |
| BTC-5 |
HMCN1 |
83872 |
1 |
186057081 |
186057084 |
Frame_Shift_Del |
GTAT |
- |
c.9381_9384delGT |
c.(9379-93p.QY3127fs |
hemicentin 1 |
| BTC-5 |
UPP2 |
151531 |
2 |
158977957 |
158977958 |
Frame_Shift_Ins |
- |
T |
c.491_492insT |
c.(490-495)p.V165fs |
uridine phosphorylase 2 |
| BTC-5 |
FNTB |
2342 |
14 |
65511053 |
65511053 |
Nonsense_Mutation |
C |
T |
c.847C>T |
c.(847-849)p.R283* |
farnesyltransferase, CAAX box, beta |
| BTC-5 |
ACSM2A |
123876 |
16 |
20482871 |
20482871 |
Nonsense_Mutation |
C |
T |
c.754C>T |
c.(754-756)p.Q252* |
acyl-CoA synthetase medium-chain family member 2A |
| BTC-5 |
PSD3 |
23362 |
8 |
18430175 |
18430175 |
Nonsense_Mutation |
C |
A |
c.2647G>T |
c.(2647-264p.E883* |
pleckstrin and Sec7 domain containing 3 |
| BTC-5 |
COL11A1 |
1301 |
1 |
103467527 |
103467527 |
Splice_Site |
T |
C |
|
c.e24-2 |
collagen, type XI, alpha 1 |
| BTC-5 |
AC007405. |
0 |
2 |
171570595 |
171570595 |
Missense_Mutation |
G |
C |
c.359C>G |
c.(358-360)p.A120G |
|
| BTC-5 |
AC009365. |
0 |
7 |
132412557 |
132412557 |
Missense_Mutation |
G |
A |
c.409G>A |
c.(409-411)p.V137I |
|
| BTC-5 |
ADAM21 |
8747 |
14 |
70924614 |
70924614 |
Missense_Mutation |
G |
T |
c.398G>T |
c.(397-399)p.R133L |
ADAM metallopeptidase domain 21 |
| BTC-5 |
APOOL |
139322X |
|
84329077 |
84329077 |
Missense_Mutation |
G |
C |
c.560G>C |
c.(559-561)p.S187T |
apolipoprotein O-like |
| BTC-5 |
ASPM |
259266 |
1 |
197074062 |
197074062 |
Missense_Mutation |
T |
C |
c.4319A>G |
c.(4318-43p.Q1440R |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| BTC-5 |
ATP7A |
538X |
|
77301051 |
77301051 |
Missense_Mutation |
C |
A |
c.4208C>A |
c.(4207-42p.S1403Y |
ATPase, Cu++ transporting, alpha polypeptide |
| BTC-5 |
ATP8B4 |
79895 |
15 |
50288883 |
50288883 |
Missense_Mutation |
C |
G |
c.580G>C |
c.(580-582)p.G194R |
ATPase, class I, type 8B, member 4 |
| BTC-5 |
CACNA1A |
773 |
19 |
13616812 |
13616812 |
Missense_Mutation |
C |
A |
c.227G>T |
c.(226-228)p.R76L |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| BTC-5 |
CCDC175 |
729665 |
14 |
60031860 |
60031860 |
Missense_Mutation |
T |
C |
c.625A>G |
c.(625-627)p.I209V |
coiled-coil domain containing 175 |
| BTC-5 |
CCL18 |
6362 |
17 |
34398340 |
34398340 |
Missense_Mutation |
G |
A |
c.209G>A |
c.(208-210)p.C70Y |
chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) |
| BTC-5 |
CD163 |
9332 |
12 |
7651580 |
7651580 |
Missense_Mutation |
A |
G |
c.662T>C |
c.(661-663)p.I221T |
CD163 molecule |
| BTC-5 |
COL21A1 |
81578 |
6 |
55922544 |
55922544 |
Missense_Mutation |
G |
T |
c.2785C>A |
c.(2785-27p.Q929K |
collagen, type XXI, alpha 1 |
| BTC-5 |
CSMD3 |
114788 |
8 |
113697935 |
113697935 |
Missense_Mutation |
G |
C |
c.2182C>G |
c.(2182-21p.P728A |
CUB and Sushi multiple domains 3 |
| BTC-5 |
CXorf28 |
1E+08X |
|
3189922 |
3189922 |
Missense_Mutation |
C |
T |
c.49C>T |
c.(49-51)Ccp.P17S |
chromosome X open reading frame 28 |
| BTC-5 |
DMXL2 |
23312 |
15 |
51791693 |
51791693 |
Missense_Mutation |
T |
A |
c.3728A>T |
c.(3727-37p.D1243V |
Dmx-like 2 |
| BTC-5 |
GAS2 |
2620 |
11 |
22747946 |
22747946 |
Missense_Mutation |
G |
A |
c.376G>A |
c.(376-378)p.D126N |
growth arrest-specific 2 |
| BTC-5 |
GPR98 |
84059 |
5 |
89939702 |
89939702 |
Missense_Mutation |
C |
T |
c.2636C>T |
c.(2635-26p.T879M |
G protein-coupled receptor 98 |
| BTC-5 |
HOXA4 |
3201 |
7 |
27170310 |
27170310 |
Missense_Mutation |
G |
T |
c.43C>A |
c.(43-45)Ccp.P15T |
homeobox A4 |
| BTC-5 |
HP |
3240 |
16 |
72090131 |
72090131 |
Missense_Mutation |
C |
T |
c.77C>T |
c.(76-78)aCp.T26M |
haptoglobin |
| BTC-5 |
ICAM5 |
7087 |
19 |
10405259 |
10405259 |
Missense_Mutation |
T |
C |
c.2173T>C |
c.(2173-21p.C725R |
intercellular adhesion molecule 5, telencephalin |
| BTC-5 |
KIAA1244 |
57221 |
6 |
138576619 |
138576619 |
Missense_Mutation |
C |
T |
c.817C>T |
c.(817-819)p.H273Y |
KIAA1244 |
| BTC-5 |
KIF21B |
23046 |
1 |
200948693 |
200948693 |
Missense_Mutation |
C |
T |
c.4130G>A |
c.(4129-41p.R1377Q |
kinesin family member 21B |
| BTC-5 |
KRTAP21-1 |
337977 |
21 |
32127554 |
32127554 |
Missense_Mutation |
C |
A |
c.143G>T |
c.(142-144)p.C48F |
keratin associated protein 21-1 |
| BTC-5 |
LCA10 |
0X |
|
153152431 |
153152431 |
Missense_Mutation |
G |
T |
c.311G>T |
c.(310-312)p.R104L |
|
| BTC-5 |
MAP1B |
4131 |
5 |
71490888 |
71490888 |
Missense_Mutation |
C |
G |
c.1706C>G |
c.(1705-17p.T569R |
microtubule-associated protein 1B |
| BTC-5 |
MLK4 |
0 |
1 |
233518335 |
233518335 |
Missense_Mutation |
C |
T |
c.2989C>T |
c.(2989-29p.P997S |
|
| BTC-5 |
NBEAL2 |
23218 |
3 |
47041430 |
47041430 |
Missense_Mutation |
C |
A |
c.3841C>A |
c.(3841-384p.Q1281K |
neurobeachin-like 2 |
| BTC-5 |
NFATC2 |
4773 |
20 |
50140482 |
50140482 |
Missense_Mutation |
C |
T |
c.298G>A |
c.(298-300)p.A100T |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
| BTC-5 |
NRAS |
4893 |
1 |
115256529 |
115256529 |
Missense_Mutation |
T |
C |
c.182A>G |
c.(181-183)p.Q61R |
neuroblastoma RAS viral (v-ras) oncogene homolog |
| BTC-5 |
OLFML2B |
25903 |
1 |
161967939 |
161967939 |
Missense_Mutation |
T |
A |
c.1153A>T |
c.(1153-11p.S385C |
olfactomedin-like 2B |
| BTC-5 |
OSBPL5 |
114879 |
11 |
3143583 |
3143583 |
Missense_Mutation |
G |
A |
c.295C>T |
c.(295-297)p.L99F |
oxysterol binding protein-like 5 |
| BTC-5 |
PCDHB1 |
29930 |
5 |
140432385 |
140432385 |
Missense_Mutation |
G |
A |
c.1330G>A |
c.(1330-13p.D444N |
protocadherin beta 1 |
| BTC-5 |
PCSK5 |
5125 |
9 |
78936474 |
78936474 |
Missense_Mutation |
G |
A |
c.3940G>A |
c.(3940-394p.E1314K |
proprotein convertase subtilisin/kexin type 5 |
| BTC-5 |
PGR |
5241 |
11 |
100998706 |
100998706 |
Missense_Mutation |
C |
T |
c.1096G>A |
c.(1096-10p.D366N |
progesterone receptor |
| BTC-5 |
PHIP |
55023 |
6 |
79695085 |
79695085 |
Missense_Mutation |
T |
G |
c.2521A>C |
c.(2521-25p.S841R |
pleckstrin homology domain interacting protein |
| BTC-5 |
PIK3C2G |
5288 |
12 |
18435656 |
18435656 |
Missense_Mutation |
C |
A |
c.641C>A |
c.(640-642)p.P214H |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| BTC-5 |
PRKCD |
5580 |
3 |
53219710 |
53219710 |
Missense_Mutation |
A |
T |
c.979A>T |
c.(979-981)p.M327L |
protein kinase C, delta |
| BTC-5 |
SEC14L2 |
23541 |
22 |
30802345 |
30802345 |
Missense_Mutation |
C |
G |
c.145C>G |
c.(145-147)p.L49V |
SEC14-like 2 (S. cerevisiae) |
| BTC-5 |
SEMA6A |
57556 |
5 |
115782872 |
115782872 |
Missense_Mutation |
C |
T |
c.2530G>A |
c.(2530-25p.E844K |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| BTC-5 |
SKIV2L2 |
23517 |
5 |
54635888 |
54635888 |
Missense_Mutation |
G |
T |
c.566G>T |
c.(565-567)p.S189I |
superkiller viralicidic activity 2-like 2 (S. cerevisiae) |
| BTC-5 |
TAF1L |
138474 |
9 |
32631848 |
32631848 |
Missense_Mutation |
G |
A |
c.3730C>T |
c.(3730-37p.R1244W |
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 210kDa-like |
| BTC-5 |
TEX15 |
56154 |
8 |
30702546 |
30702546 |
Missense_Mutation |
T |
G |
c.3988A>C |
c.(3988-39p.K1330Q |
testis expressed 15 |
| BTC-5 |
TLR8 |
51311X |
|
12939230 |
12939230 |
Missense_Mutation |
G |
C |
c.2125G>C |
c.(2125-21p.E709Q |
toll-like receptor 8 |
| BTC-5 |
TMEM200A |
114801 |
6 |
130762039 |
130762039 |
Missense_Mutation |
A |
G |
c.472A>G |
c.(472-474)p.I158V |
transmembrane protein 200A |
| BTC-5 |
TMEM80 |
283232 |
11 |
703112 |
703112 |
Missense_Mutation |
G |
T |
c.469G>T |
c.(469-471)p.V157F |
transmembrane protein 80 |
| BTC-5 |
TRPM6 |
140803 |
9 |
77377993 |
77377993 |
Missense_Mutation |
C |
A |
c.3594G>T |
c.(3592-35p.K1198N |
transient receptor potential cation channel, subfamily M, member 6 |
| BTC-5 |
TSPYL5 |
85453 |
8 |
98290039 |
98290039 |
Missense_Mutation |
G |
A |
c.34C>T |
c.(34-36)Cgp.R12C |
TSPY-like 5 |
| BTC-6 |
AMIGO2 |
347902 |
12 |
47471598 |
47471598 |
Missense_Mutation |
A |
C |
c.1188T>G |
c.(1186-11p.F396L |
adhesion molecule with Ig-like domain 2 |
| BTC-6 |
BIRC6 |
57448 |
2 |
32712788 |
32712788 |
Missense_Mutation |
A |
T |
c.7888A>T |
c.(7888-78p.I2630F |
baculoviral IAP repeat containing 6 |
| BTC-6 |
BZW1 |
9689 |
2 |
201680210 |
201680210 |
Missense_Mutation |
T |
G |
c.211T>G |
c.(211-213)p.F71V |
basic leucine zipper and W2 domains 1 |
| BTC-6 |
CD200R1 |
131450 |
3 |
112648303 |
112648303 |
Missense_Mutation |
G |
A |
c.185C>T |
c.(184-186)p.P62L |
CD200 receptor 1 |
| BTC-6 |
CRYBG3 |
131544 |
3 |
97593426 |
97593426 |
Missense_Mutation |
G |
T |
c.3388G>T |
c.(3388-33p.G1130W |
beta-gamma crystallin domain containing 3 |
| BTC-6 |
DEAF1 |
10522 |
11 |
694992 |
694992 |
Missense_Mutation |
A |
G |
c.56T>C |
c.(55-57)gTp.V19A |
DEAF1 transcription factor |
| BTC-6 |
EFCAB5 |
374786 |
17 |
28295933 |
28295933 |
Missense_Mutation |
A |
T |
c.315A>T |
c.(313-315)p.E105D |
EF-hand calcium binding domain 5 |
| BTC-6 |
FSIP2 |
401024 |
2 |
186671383 |
186671383 |
Missense_Mutation |
G |
A |
c.17350G>A |
c.(17350-1p.A5784T |
fibrous sheath interacting protein 2 |
| BTC-6 |
GAPVD1 |
26130 |
9 |
128064438 |
128064438 |
Missense_Mutation |
A |
T |
c.362A>T |
c.(361-363)p.N121I |
GTPase activating protein and VPS9 domains 1 |
| BTC-6 |
IQSEC2 |
23096X |
|
53349985 |
53349985 |
Missense_Mutation |
C |
T |
c.337G>A |
c.(337-339)p.D113N |
IQ motif and Sec7 domain 2 |
| BTC-6 |
KDM5A |
5927 |
12 |
427378 |
427378 |
Missense_Mutation |
C |
T |
c.2791G>A |
c.(2791-27p.G931R |
lysine (K)-specific demethylase 5A |
| BTC-6 |
LPAR3 |
23566 |
1 |
85279695 |
85279695 |
Missense_Mutation |
T |
C |
c.896A>G |
c.(895-897)p.D299G |
lysophosphatidic acid receptor 3 |
| BTC-6 |
NLRP1 |
22861 |
17 |
5462767 |
5462767 |
Missense_Mutation |
A |
G |
c.1249T>C |
c.(1249-12p.W417R |
NLR family, pyrin domain containing 1 |
| BTC-6 |
OR6B3 |
150681 |
2 |
240984973 |
240984973 |
Missense_Mutation |
C |
A |
c.517G>T |
c.(517-519)p.V173F |
olfactory receptor, family 6, subfamily B, member 3 |
| BTC-6 |
OTUD6A |
139562X |
|
69282627 |
69282627 |
Missense_Mutation |
G |
C |
c.253G>C |
c.(253-255)p.E85Q |
OTU deubiquitinase 6A |
| BTC-6 |
PTPRH |
5794 |
19 |
55715264 |
55715264 |
Missense_Mutation |
T |
C |
c.772A>G |
c.(772-774)p.T258A |
protein tyrosine phosphatase, receptor type, H |
| BTC-6 |
RAD50 |
10111 |
5 |
131927660 |
131927660 |
Missense_Mutation |
T |
G |
c.1727T>G |
c.(1726-17p.L576R |
RAD50 homolog (S. cerevisiae) |
| BTC-6 |
RB1 |
5925 |
13 |
49039417 |
49039417 |
Missense_Mutation |
G |
C |
c.2402G>C |
c.(2401-24p.G801A |
retinoblastoma 1 |
| BTC-6 |
SAMD9L |
219285 |
7 |
92763376 |
92763376 |
Missense_Mutation |
C |
T |
c.1909G>A |
c.(1909-19p.A637T |
sterile alpha motif domain containing 9-like |
| BTC-6 |
SERINC5 |
256987 |
5 |
79454696 |
79454696 |
Missense_Mutation |
C |
G |
c.949G>C |
c.(949-951)p.G317R |
serine incorporator 5 |
| BTC-6 |
ST6GALNA |
81849 |
1 |
77515950 |
77515950 |
Missense_Mutation |
T |
A |
c.679T>A |
c.(679-681)p.S227T |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| BTC-6 |
TMEM47 |
83604X |
|
34657486 |
34657486 |
Missense_Mutation |
A |
G |
c.245T>C |
c.(244-246)p.L82P |
transmembrane protein 47 |
| BTC-6 |
TRIM41 |
90933 |
5 |
180651595 |
180651595 |
Missense_Mutation |
T |
C |
c.596T>C |
c.(595-597)p.L199P |
tripartite motif containing 41 |
| BTC-6 |
WDSUB1 |
151525 |
2 |
160139525 |
160139525 |
Missense_Mutation |
G |
A |
c.56C>T |
c.(55-57)gCp.A19V |
WD repeat, sterile alpha motif and U-box domain containing 1 |
| BTC-6 |
ZC3H13 |
23091 |
13 |
46616352 |
46616352 |
Missense_Mutation |
C |
T |
c.286G>A |
c.(286-288)p.V96M |
zinc finger CCCH-type containing 13 |
| BTC-7 |
CCDC18 |
343099 |
1 |
93705341 |
93705341 |
Nonsense_Mutation |
G |
T |
c.2866G>T |
c.(2866-28p.E956* |
coiled-coil domain containing 18 |
| BTC-7 |
KANSL1 |
284058 |
17 |
44115975 |
44115975 |
Nonsense_Mutation |
G |
A |
c.2281C>T |
c.(2281-22p.R761* |
KAT8 regulatory NSL complex subunit 1 |
| BTC-7 |
PLEKHG2 |
64857 |
19 |
39912773 |
39912773 |
Nonsense_Mutation |
G |
T |
c.1522G>T |
c.(1522-15p.E508* |
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| BTC-7 |
ZNF546 |
339327 |
19 |
40520465 |
40520465 |
Nonsense_Mutation |
C |
T |
c.1288C>T |
c.(1288-12p.R430* |
zinc finger protein 546 |
| BTC-7 |
SI |
6476 |
3 |
164760855 |
164760855 |
Nonsense_Mutation |
C |
A |
c.1996G>T |
c.(1996-19p.G666* |
sucrase-isomaltase (alpha-glucosidase) |
| BTC-7 |
SLITRK2 |
84631X |
|
144904042 |
144904042 |
Nonsense_Mutation |
T |
A |
c.99T>A |
c.(97-99)tgp.C33* |
SLIT and NTRK-like family, member 2 |
| BTC-7 |
CNTNAP5 |
129684 |
2 |
125367471 |
125367471 |
Missense_Mutation |
C |
T |
c.1847C>T |
c.(1846-184p.P616L |
contactin associated protein-like 5 |
| BTC-7 |
DNMT3A |
1788 |
2 |
25467461 |
25467461 |
Missense_Mutation |
T |
C |
c.946A>G |
c.(946-948)p.I316V |
DNA (cytosine-5-)-methyltransferase 3 alpha |
| BTC-7 |
DSG4 |
147409 |
18 |
28971077 |
28971077 |
Missense_Mutation |
G |
A |
c.721G>A |
c.(721-723)p.D241N |
desmoglein 4 |
| BTC-7 |
EPC1 |
80314 |
10 |
32560813 |
32560813 |
Missense_Mutation |
T |
C |
c.2038A>G |
c.(2038-204p.T680A |
enhancer of polycomb homolog 1 (Drosophila) |
| BTC-7 |
ESM1 |
11082 |
5 |
54277867 |
54277867 |
Missense_Mutation |
A |
T |
c.409T>A |
c.(409-411)p.Y137N |
endothelial cell-specific molecule 1 |
| BTC-7 |
F13A1 |
2162 |
6 |
6266956 |
6266956 |
Missense_Mutation |
C |
A |
c.406G>T |
c.(406-408)p.V136F |
coagulation factor XIII, A1 polypeptide |
| BTC-7 |
FCRL3 |
115352 |
1 |
157665154 |
157665154 |
Missense_Mutation |
G |
A |
c.1376C>T |
c.(1375-13p.A459V |
Fc receptor-like 3 |
| BTC-7 |
FUT1 |
2523 |
19 |
49254093 |
49254093 |
Missense_Mutation |
C |
A |
c.446G>T |
c.(445-447)p.W149L |
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) |
| BTC-7 |
GLI3 |
2737 |
7 |
42005951 |
42005951 |
Missense_Mutation |
C |
A |
c.2720G>T |
c.(2719-27p.S907I |
GLI family zinc finger 3 |
| BTC-7 |
HEATR1 |
55127 |
1 |
236721708 |
236721708 |
Missense_Mutation |
C |
T |
c.5033G>A |
c.(5032-50p.C1678Y |
HEAT repeat containing 1 |
| BTC-7 |
KLHL24 |
54800 |
3 |
183368493 |
183368493 |
Missense_Mutation |
A |
G |
c.349A>G |
c.(349-351)p.M117V |
kelch-like family member 24 |
| BTC-7 |
KLHL9 |
55958 |
9 |
21333179 |
21333179 |
Missense_Mutation |
C |
G |
c.1680G>C |
c.(1678-16p.M560I |
kelch-like family member 9 |
| BTC-7 |
KMT2A |
4297 |
11 |
118392628 |
118392628 |
Missense_Mutation |
T |
A |
c.11660T>A |
c.(11659-1p.M3887K |
lysine (K)-specific methyltransferase 2A |
| BTC-7 |
KRAS |
3845 |
12 |
25398284 |
25398284 |
Missense_Mutation |
C |
A |
c.35G>T |
c.(34-36)gGp.G12V |
Kirsten rat sarcoma viral oncogene homolog |
| BTC-7 |
LPP |
4026 |
3 |
188326976 |
188326976 |
Missense_Mutation |
C |
T |
c.457C>T |
c.(457-459)p.P153S |
LIM domain containing preferred translocation partner in lipoma |
| BTC-7 |
LRP1B |
53353 |
2 |
141707920 |
141707920 |
Missense_Mutation |
A |
G |
c.3020T>C |
c.(3019-30p.F1007S |
low density lipoprotein receptor-related protein 1B |
| BTC-7 |
MSH3 |
4437 |
5 |
79950739 |
79950739 |
Missense_Mutation |
G |
A |
c.193G>A |
c.(193-195)p.A65T |
mutS homolog 3 |
| BTC-7 |
NCOR1 |
9611 |
17 |
16049747 |
16049747 |
Missense_Mutation |
T |
C |
c.1025A>G |
c.(1024-10p.Y342C |
nuclear receptor corepressor 1 |
| BTC-7 |
NIPBL |
25836 |
5 |
37064917 |
37064917 |
Missense_Mutation |
A |
G |
c.8338A>G |
c.(8338-834p.S2780G |
Nipped-B homolog (Drosophila) |
| BTC-7 |
NTRK1 |
4914 |
1 |
156851340 |
156851340 |
Missense_Mutation |
G |
A |
c.2297G>A |
c.(2296-22p.R766Q |
neurotrophic tyrosine kinase, receptor, type 1 |
| BTC-7 |
PELP1 |
27043 |
17 |
4575146 |
4575146 |
Missense_Mutation |
C |
T |
c.3140G>A |
c.(3139-314p.G1047E |
proline, glutamate and leucine rich protein 1 |
| BTC-7 |
RELN |
5649 |
7 |
103183183 |
103183183 |
Missense_Mutation |
A |
T |
c.6666T>A |
c.(6664-66p.H2222Q |
reelin |
| BTC-7 |
SPTBN1 |
6711 |
2 |
54871578 |
54871578 |
Missense_Mutation |
G |
A |
c.4124G>A |
c.(4123-41p.R1375Q |
spectrin, beta, non-erythrocytic 1 |
| BTC-7 |
SUSD2 |
56241 |
22 |
24583685 |
24583685 |
Missense_Mutation |
G |
T |
c.2038G>T |
c.(2038-204p.A680S |
sushi domain containing 2 |
| BTC-7 |
ZNF14 |
7561 |
19 |
19822422 |
19822422 |
Missense_Mutation |
C |
A |
c.1668G>T |
c.(1666-16p.E556D |
zinc finger protein 14 |
| BTC-8 |
APC |
324 |
5 |
112175952 |
112175952 |
Frame_Shift_Del |
A |
- |
c.4661delA |
c.(4660-46p.E1554fs |
adenomatous polyposis coli |
| BTC-8 |
LMO7 |
4008 |
13 |
76408475 |
76408475 |
Nonsense_Mutation |
C |
T |
c.2479C>T |
c.(2479-24p.Q827* |
LIM domain 7 |
| BTC-8 |
TP53 |
7157 |
17 |
7574003 |
7574003 |
Nonsense_Mutation |
G |
A |
c.1024C>T |
c.(1024-10p.R342* |
tumor protein p53 |
| BTC-8 |
DUXA |
503835 |
19 |
57670637 |
57670637 |
Nonsense_Mutation |
G |
A |
c.190C>T |
c.(190-192)p.Q64* |
double homeobox A |
| BTC-8 |
MMS22L |
253714 |
6 |
97676947 |
97676947 |
Nonsense_Mutation |
C |
T |
c.1862G>A |
c.(1861-18p.W621* |
MMS22-like, DNA repair protein |
| BTC-8 |
KIF15 |
56992 |
3 |
44843467 |
44843467 |
Splice_Site |
G |
C |
|
c.e13+1 |
kinesin family member 15 |
| BTC-8 |
ABCA9 |
10350 |
17 |
67025361 |
67025361 |
Missense_Mutation |
T |
G |
c.1453A>C |
c.(1453-14p.N485H |
ATP-binding cassette, sub-family A (ABC1), member 9 |
| BTC-8 |
ACSS1 |
84532 |
20 |
25004233 |
25004233 |
Missense_Mutation |
C |
T |
c.676G>A |
c.(676-678)p.G226R |
acyl-CoA synthetase short-chain family member 1 |
| BTC-8 |
ADAMTS6 |
11174 |
5 |
64595872 |
64595872 |
Missense_Mutation |
G |
A |
c.1310C>T |
c.(1309-13p.A437V |
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| BTC-8 |
ADORA1 |
134 |
1 |
203098180 |
203098180 |
Missense_Mutation |
A |
G |
c.211A>G |
c.(211-213)p.I71V |
adenosine A1 receptor |
| BTC-8 |
AOC3 |
8639 |
17 |
41006705 |
41006705 |
Missense_Mutation |
C |
T |
c.1841C>T |
c.(1840-184p.P614L |
amine oxidase, copper containing 3 |
| BTC-8 |
CPZ |
8532 |
4 |
8603105 |
8603105 |
Missense_Mutation |
G |
A |
c.344G>A |
c.(343-345)p.R115Q |
carboxypeptidase Z |
| BTC-8 |
DACT3 |
147906 |
19 |
47151783 |
47151783 |
Missense_Mutation |
G |
C |
c.1846C>G |
c.(1846-184p.L616V |
dishevelled-binding antagonist of beta-catenin 3 |
| BTC-8 |
ENPP2 |
5168 |
8 |
120577135 |
120577135 |
Missense_Mutation |
A |
C |
c.1097T>G |
c.(1096-10p.V366G |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| BTC-8 |
INO80D |
54891 |
2 |
206870153 |
206870153 |
Missense_Mutation |
C |
A |
c.2023G>T |
c.(2023-20p.A675S |
INO80 complex subunit D |
| BTC-8 |
IRX3 |
79191 |
16 |
54319378 |
54319378 |
Missense_Mutation |
T |
C |
c.415A>G |
c.(415-417)p.T139A |
iroquois homeobox 3 |
| BTC-8 |
KRAS |
3845 |
12 |
25398281 |
25398281 |
Missense_Mutation |
C |
T |
c.38G>A |
c.(37-39)gGp.G13D |
Kirsten rat sarcoma viral oncogene homolog |
| BTC-8 |
KRT73 |
319101 |
12 |
53004589 |
53004589 |
Missense_Mutation |
C |
T |
c.1141G>A |
c.(1141-114p.A381T |
keratin 73 |
| BTC-8 |
LAX1 |
54900 |
1 |
203743063 |
203743063 |
Missense_Mutation |
G |
A |
c.451G>A |
c.(451-453)p.A151T |
lymphocyte transmembrane adaptor 1 |
| BTC-8 |
MATN4 |
8785 |
20 |
43927089 |
43927089 |
Missense_Mutation |
G |
A |
c.1147C>T |
c.(1147-114p.R383W |
matrilin 4 |
| BTC-8 |
NAB1 |
4664 |
2 |
191524551 |
191524551 |
Missense_Mutation |
G |
A |
c.649G>A |
c.(649-651)p.E217K |
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| BTC-8 |
NPR3 |
4883 |
5 |
32712527 |
32712527 |
Missense_Mutation |
C |
A |
c.645C>A |
c.(643-645)p.F215L |
natriuretic peptide receptor 3 |
| BTC-8 |
OTOG |
340990 |
11 |
17655819 |
17655819 |
Missense_Mutation |
A |
G |
c.7288A>G |
c.(7288-72p.T2430A |
otogelin |
| BTC-8 |
PCDHGB6 |
56100 |
5 |
140788665 |
140788665 |
Missense_Mutation |
G |
A |
c.896G>A |
c.(895-897)p.G299D |
protocadherin gamma subfamily B, 6 |
| BTC-8 |
POLA1 |
5422X |
|
24906154 |
24906154 |
Missense_Mutation |
G |
A |
c.4061G>A |
c.(4060-40p.R1354H |
polymerase (DNA directed), alpha 1, catalytic subunit |
| BTC-8 |
RALGAPA1 |
253959 |
14 |
36096402 |
36096402 |
Missense_Mutation |
T |
A |
c.5272A>T |
c.(5272-52p.M1758L |
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| BTC-8 |
RIPK2 |
8767 |
8 |
90792331 |
90792331 |
Missense_Mutation |
G |
C |
c.882G>C |
c.(880-882)p.L294F |
receptor-interacting serine-threonine kinase 2 |
| BTC-8 |
ROBO1 |
6091 |
3 |
78710287 |
78710287 |
Missense_Mutation |
A |
G |
c.2213T>C |
c.(2212-22p.I738T |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| BTC-8 |
ROBO2 |
6092 |
3 |
77657029 |
77657029 |
Missense_Mutation |
C |
A |
c.3217C>A |
c.(3217-32p.P1073T |
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| BTC-8 |
SHISA8 |
440829 |
22 |
42310285 |
42310285 |
Missense_Mutation |
C |
A |
c.286G>T |
c.(286-288)p.A96S |
shisa family member 8 |
| BTC-8 |
SMARCA2 |
6595 |
9 |
2104152 |
2104152 |
Missense_Mutation |
T |
C |
c.3275T>C |
c.(3274-32p.L1092P |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| BTC-8 |
SSTR1 |
6751 |
14 |
38678956 |
38678956 |
Missense_Mutation |
G |
A |
c.362G>A |
c.(361-363)p.R121H |
somatostatin receptor 1 |
| BTC-8 |
SYN3 |
8224 |
22 |
33265027 |
33265027 |
Missense_Mutation |
C |
T |
c.547G>A |
c.(547-549)p.G183S |
synapsin III |
| BTC-8 |
TROVE2 |
6738 |
1 |
193046115 |
193046115 |
Missense_Mutation |
C |
G |
c.1021C>G |
c.(1021-10p.L341V |
TROVE domain family, member 2 |
| BTC-8 |
USH2A |
7399 |
1 |
215844423 |
215844423 |
Missense_Mutation |
C |
T |
c.14024G>A |
c.(14023-14p.R4675K |
Usher syndrome 2A (autosomal recessive, mild) |
| BTC-8 |
USH2A |
7399 |
1 |
216062042 |
216062042 |
Missense_Mutation |
G |
C |
c.7949C>G |
c.(7948-79p.P2650R |
Usher syndrome 2A (autosomal recessive, mild) |
| BTC-8 |
XCR1 |
2829 |
3 |
46062623 |
46062623 |
Missense_Mutation |
G |
A |
c.817C>T |
c.(817-819)p.R273C |
chemokine (C motif) receptor 1 |
| BTC-8 |
ZSCAN20 |
7579 |
1 |
33960845 |
33960845 |
Missense_Mutation |
C |
A |
c.2901C>A |
c.(2899-29p.F967L |
zinc finger and SCAN domain containing 20 |