|
 |
- Search
| Genomics Inform > Volume 20(4); 2022 > Article |
|
Abstract
Many packages for a meta-analysis of genome-wide association studies (GWAS) have been developed to discover genetic variants. Although variations across studies must be considered, there are not many currently-accessible packages that estimate between-study heterogeneity. Thus, we propose a python based application called Beta-Meta which can easily process a meta-analysis by automatically selecting between a fixed effects and a random effects model based on heterogeneity. Beta-Meta implements flexible input data manipulation to allow multiple meta-analyses of different genotype-phenotype associations in a single process. It provides a step-by-step meta-analysis of GWAS for each association in the following order: heterogeneity test, two different calculations of an effect size and a p-value based on heterogeneity, and the Benjamini-Hochberg p-value adjustment. These methods enable users to validate the results of individual studies with greater statistical power and better estimation precision. We elaborate on these and illustrate them with examples from several studies of infertility-related disorders.
Genome-wide association studies (GWAS) of diseases and traits have increasingly been used to identify single nucleotide polymorphisms (SNPs). Although GWAS have tested hundreds of thousands of genetic variants to discover genotype-phenotype associations, they have a few limitations. Variants discovered in individual GWAS explain only a small proportion of heritability, and their genetic effect sizes are mostly small and require a substantial sample size to identify [1,2]. Moreover, some studies examining the same genotype-phenotype association yield inconsistent results such as variant effect sizes in opposite directions [3,4]. To overcome these limitations, a meta-analysis of GWAS has been used extensively since it can improve the statistical power by combining data across any number of independent studies and can clarify heterogeneity among their results [5].
As meta-analysis has become a popular tool for aggregating data from multiple sources, several studies have revised analytical strategies from previous well-known studies [6-9]. A weighted average of the effect sizes can be calculated under a fixed effects model or a random effects model, but the fixed effects model can lead to false-positive results when there is heterogeneity between studies [9,10]. Even though it is important to use the appropriate approaches for meta-analyses, there are few available tools that provide a step-by-step calculation, running both the fixed effects model and the random effects model [10]. Therefore, for those who find it difficult to conduct a meta-analysis, we have developed a flexible data processing tool that adopts the revised methods assessing heterogeneity between studies and using the Benjamini-Hochberg (BH) procedure to calculate adjusted p-values [11]. In addition to these methods, Beta-Meta has several convenient features such as an automatic selection between the two models depending upon the quantified heterogeneity. It also manifests flexibility and convenience in processing data as it can perform a varying number of meta-analyses simultaneously and operate strand flipping automatically when there is a discrepancy in the direction of the strand orientation between studies. Also, we have attached haploR package [12] which detects alternative SNPs by estimating their correlations.
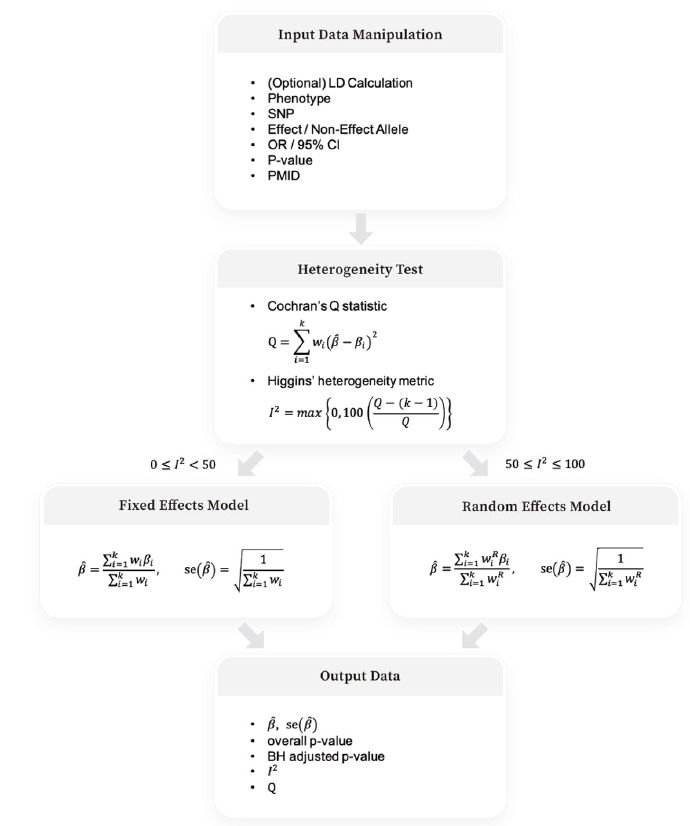
Fig. 1 depicts the four steps of Beta-Meta: input data manipulation, heterogeneity test, weighted effect size calculation under the fixed and random effects models, and output data of summary statistics after the BH adjustment.
Meta-analysis can improve signal detection when we account for not only between-study heterogeneity but also differences in linkage disequilibrium (LD) between ethnicities [35]; in addition, several trans-ethnic meta-analyses have identified unknown susceptibility genes [35-37]. As it is important to consider differences in LD, we utilize the haploR package [12] that queries HaploReg database [38] and returns alternative SNPs in LD. By calculating pairwise metrics of LD in each continental population, LD structures between ethnicities can be discovered and hence alternative SNPs can be used for the following meta-analysis [38]. This step is optional; users may skip this step and start a meta-analysis when the summary statistics of their target SNPs of interest are already obtained.
After surveying the studies of interest (infertility-related disorders in this paper), we created a table for input data in Excel (Supplementary Table 1). Beta-Meta can read an Excel file for input data, which must include phenotypes, SNPs, effect and non-effect alleles, effect sizes, and p-values. For the effect sizes and their levels of significance, either the beta coefficient and its standard error or the odds ratio (OR) and its confidence interval can be used. As Beta-Meta calculates SNP-phenotype associations separately, it is acceptable to include as many phenotypes as desired in the single input file.
When the OR and its confidence interval are used for input data, they are converted into the beta coefficient and the standard error, respectively. The normalized effect of the ithstudy, βi is the logarithm of OR, where k is the number of individual studies, each of which is designed to examine the same SNP-phenotype association [9].
The standard error si is calculated from the 95% confidence interval of the OR.
When synthesizing datasets for meta-analysis, it is important to ensure uniformity in allele labels and hence in the direction of the effect because alleles are typically called on only one of the two DNA strands in sequencing experiments [39]. Beta-Meta automatically corrects the direction of the effect by using one of the datasets with the lowest p-value as a reference and aligning the other datasets to it. For example, when the effect and the non-effect allele are inverted between the independent studies (e.g., rs13405782 and rs1801133 as shown in Table 1), this can be resolved automatically by changing the sign of the normalized effect.
In meta-analysis, datasets generated by multiple groups by different methods are likely to have any kind of variability, also known as heterogeneity. Heterogeneity indicates that the observed effects in datasets are more different from each other than would be expected by random error alone [40]. To check the heterogeneity, the weighted average of the effect size
β ^
Then, we calculate the Cochran’s Q statistic, Q and Higgins’ heterogeneity metric, I2 for the heterogeneity test [6].
I2 quantifies the degree of heterogeneity as a value between 0 and 100% [41]. As a greater value of I2 indicates stronger heterogeneity, the weighted average of the effect sizes is calculated, based on I2, using two different models: the fixed effects model and the random effects model. A threshold value of I2 for the model selection is set to 50%.
For 0≤I2<50, we use the fixed effects model to calculate the weighted average of the effect sizes and its standard error [7].
The integrated p-value through meta-analysis can be obtained as follows [7]:
where Φ is the cumulative distribution function of the standard normal distribution, and integrated Z-score, Z [7] is
Finally, to reduce the false-positive results, the integrated p-values are corrected by the BH adjustment method. When p(1), p(2), ⋯,p(m) are the p-values of the SNPs sorted in ascending order (p(1) ≤ p(2) ≤ ⋯ ≤ p(m)), the adjusted p-values obtained through the BH procedure are as follows [11]:
where m is the number of different SNPs related to a specific phenotype, and j is the ranking in the ascending order of the p-values of SNPs related to the specific phenotype.
Using Beta-Meta, we performed a sample test of integrating multiple studies of infertility and obtained a table containing all of the above calculated summary statistics values (Supplementary Table 2) and a forest plot of combined effect sizes (Supplementary Fig. 1). The conventional genome-wide significance p-value threshold of 5 × 10-8 was used to identify significant SNP markers. Of the total 26 SNP-phenotype associations from the 23 studies we investigated (Supplementary Table 1), the only significant association was the one between rs10965232 and endometriosis from Uno et al. [14] with a p-value of 5.57 × 10-12 (Table 1). After performing the meta-analysis, we found three more significantly associated SNPs: rs13405728, rs1801133, and rs10842262 as displayed in Table 2.
In order to check the accuracy of Beta-Meta, we compared the meta-analysis results of Beta-Meta (Supplementary Table 2) with those of METAL [8] (Supplementary Table 3), which is one of the most widely used meta-analysis packages but does not have a random effects option. We could confirm the accuracy of Beta-Meta calculation with the result that the significantly associated SNPs identified by METAL and those found by Beta-Meta were the same. At the same time, Beta-Meta features convenience as it calculates the summary statistics accurately by automatically selecting the appropriate model based on heterogeneity.
Beta-Meta application can be utilized as an effortless meta-analysis tool for researchers with limited statistics backgrounds. It allows them to easily manipulate and analyze their own datasets on a personal computer as it is written in python and can be run with an executable file in MS Windows.
As shown above, Beta-Meta increases the power to detect weak signals, identifying significant variants which was not significantly associated in single studies. Furthermore, it calculates the effect sizes and the p-values accurately by selecting the appropriate model based on heterogeneity and applying the BH adjustment. These can contribute to time-efficient management of the recent growth in aggregated GWAS especially for those involved in the field of genetic testing. Because it is difficult to obtain a large number of datasets and validate genotype-phenotype associations experimentally within a limited budget, meta-analysis is still in demand to discover SNP markers for genetic testing.
In conclusion, the application presented here provides a conventional and yet convenient way to conduct a meta-analysis of GWAS. Beta-Meta is expected to facilitate various research projects, such as the discovery of novel SNP markers, the calculation of polygenic risk scores, and the acquisition of biological insights into complex diseases and traits.
Notes
Authors’ Contribution
Conceptualization: WL. Data curation: GK. Formal analysis: GK. Methodology: WL, GK. Software: GK. Supervision: DK. Visualization: JHP, GK. Writing – original draft: YL, GK, WL. Writing – review & editing: DK.
Availability
Beta-Meta is written in python 3.9.7, and is available at https://github.com/Gyungbu/beta_meta.
Acknowledgments
This work was supported by National IT Industry Promotion Agency (NIPA) grant funded by the Korea government (MSIT) (No. S0252-21-1001, Development of AI Precision Medical Solution (Doctor Answer 2.0)).
Supplementary Materials
Supplementary data can be found with this article online at http://www.genominfo.org.
Supplementary Table 1.
Input data: summary statistics of the individual GWAS of infertility
Supplementary Table 3.
Output data: summary statistics after meta-analysis using METAL
Supplementary Fig. 1.
Forest plot of the combined effect sizes. Forest plot of 95% confidence intervals of the combined effect sizes after meta-analysis.
Table 1.
Example of input data: summary statistics of the individual GWAS of infertility
Table 2.
Example of output data presenting only the significantly associated SNPs after meta-analysis
| Phenotype | SNP | EA | NEA |
|
se(
|
p-value | adjusted p-value | I2 | Q |
|---|---|---|---|---|---|---|---|---|---|
| Endometriosis | rs10965235 | C | A | 0.371 | 0.046 | 8.2E-16 | 8.2E-16 | 0 | 0.083 |
| Polycystic ovary syndrome | rs13405728a | A | G | 0.371 | 0.056 | 3.55E-11 | 7.11E-11 | 71.70 | 3.534 |
| Folic acid metabolism-related male infertility | rs1801133 | C | T | -0.351 | 0.031 | 4E-29 | 8E-29 | 0 | 0.361 |
| Non-obstructive azoospermia | rs10842262 | G | C | 0.214 | 0.028 | 1.36E-14 | 1.36E-14 | 0 | 0.679 |
References
2. Tam V, Patel N, Turcotte M, Bosse Y, Pare G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet 2019;20:467–484.



3. Hu Z, Xia Y, Guo X, Dai J, Li H, Hu H, et al. A genome-wide association study in Chinese men identifies three risk loci for non-obstructive azoospermia. Nat Genet 2011;44:183–186.



4. Gu X, Li H, Chen X, Zhang X, Mei F, Jia M, et al. PEX10, SIRPA-SIRPG, and SOX5 gene polymorphisms are strongly associated with nonobstructive azoospermia susceptibility. J Assist Reprod Genet 2019;36:759–768.




5. Zeggini E, Ioannidis JP. Meta-analysis in genome-wide association studies. Pharmacogenomics 2009;10:191–201.



6. Hoaglin DC. Misunderstandings about Q and 'Cochran's Q test' in meta-analysis. Stat Med 2016;35:485–495.


7. Lee CH, Cook S, Lee JS, Han B. Comparison of two meta-analysis methods: inverse-variance-weighted average and weighted sum of Z-scores. Genomics Inform 2016;14:173–180.




8. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191.



9. Evangelou E, Ioannidis JP. Meta-analysis methods for genome-wide association studies and beyond. Nat Rev Genet 2013;14:379–389.



10. Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: a review of statistical methods and recommendations for their application. Am J Hum Genet 2010;86:6–22.



11. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995;57:289–300.


12. Zhbannikov IY, Arbeev K, Ukraintseva S, Yashin AI. haploR: an R package for querying web-based annotation tools. F1000Res 2017;6:97.




13. Lee GH, Choi YM, Hong MA, Yoon SH, Kim JJ, Hwang K, et al. Association of CDKN2B-AS and WNT4 genetic polymorphisms in Korean patients with endometriosis. Fertil Steril 2014;102:1393–1397.


14. Uno S, Zembutsu H, Hirasawa A, Takahashi A, Kubo M, Akahane T, et al. A genome-wide association study identifies genetic variants in the CDKN2BAS locus associated with endometriosis in Japanese. Nat Genet 2010;42:707–710.



15. Kim JJ, Choi YM, Hong MA, Chae SJ, Hwang K, Yoon SH, et al. FSH receptor gene p. Thr307Ala and p. Asn680Ser polymorphisms are associated with the risk of polycystic ovary syndrome. J Assist Reprod Genet 2017;34:1087–1093.




16. Wan P, Meng L, Huang C, Dai B, Jin Y, Chai L, et al. Replication study and meta-analysis of selected genetic variants and polycystic ovary syndrome susceptibility in Asian population. J Assist Reprod Genet 2021;38:2781–2789.




17. Zou J, Wu D, Liu Y, Tan S. Association of luteinizing hormone/choriogonadotropin receptor gene polymorphisms with polycystic ovary syndrome risk: a meta-analysis. Gynecol Endocrinol 2019;35:81–85.


18. Shin SJ, Lee HH, Cha SH, Kim JH, Shim SH, Choi DH, et al. Endothelial nitric oxide synthase gene polymorphisms (-786T>C, 4a4b, 894G>T) and haplotypes in Korean patients with recurrent spontaneous abortion. Eur J Obstet Gynecol Reprod Biol 2010;152:64–67.


19. Zhao X, Li Q, Yu F, Lin L, Yin W, Li J, et al. Gene polymorphism associated with endothelial nitric oxide synthase (4VNTR, G894T, C786T) and unexplained recurrent spontaneous abortion risk: a meta-analysis. Medicine (Baltimore) 2019;98:e14175.


20. Jeon YJ, Choi YS, Rah H, Kim SY, Choi DH, Cha SH, et al. Association study of microRNA polymorphisms with risk of idiopathic recurrent spontaneous abortion in Korean women. Gene 2012;494:168–173.


21. Jeon YJ, Kim SY, Rah H, Choi DH, Cha SH, Yoon TK, et al. Association of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with risk of spontaneously aborted fetuses. Am J Reprod Immunol 2012;68:408–417.


22. Sun Y, Chen M, Mao B, Cheng X, Zhang X, Xu C. Association between vascular endothelial growth factor polymorphism and recurrent pregnancy loss: a systematic review and meta-analysis. Eur J Obstet Gynecol Reprod Biol 2017;211:169–176.


23. Li L, Donghong L, Shuguang W, Hongbo Z, Jing Z, Shengbin L. Polymorphisms in the vascular endothelial growth factor gene associated with recurrent spontaneous miscarriage. J Matern Fetal Neonatal Med 2013;26:686–690.


24. Liu Z, Lin L, Yao X, Xing J. Association between polymorphisms in the XRCC1 gene and male infertility risk: a meta-analysis. Medicine (Baltimore) 2020;99:e20008.


25. Lv MQ, Li YX, Ge P, Yang YQ, Zhang J, Han SP, et al. Association between X-ray repair cross-complementing group 1 Arg399Gln polymorphism and male infertility: an update meta-analysis. Andrologia 2020;52:e13700.



26. Park JH, Lee HC, Jeong YM, Chung TG, Kim HJ, Kim NK, et al. MTHFR C677T polymorphism associates with unexplained infertile male factors. J Assist Reprod Genet 2005;22:361–368.



27. Shi TL, Wu Y, Li Y, Chen ZF, Ma YN, Zhang ZT, et al. The relevance of MTHFR C677T, A1298C, and MTRR A66G polymorphisms with response to male infertility in Asians: a meta-analysis. Medicine (Baltimore) 2019;98:e14283.


28. Lee HC, Jeong YM, Lee SH, Cha KY, Song SH, Kim NK, et al. Association study of four polymorphisms in three folate-related enzyme genes with non-obstructive male infertility. Hum Reprod 2006;21:3162–3170.


29. Chang J, Pan F, Tang Q, Wu W, Chen M, Lu C, et al. eNOS gene T786C, G894T and 4a4b polymorphisms and male infertility susceptibility: a meta-analysis. Andrologia 2017;49:e12646.


30. Song P, Zou S, Chen T, Chen J, Wang Y, Yang J, et al. Endothelial nitric oxide synthase (eNOS) T-786C, 4a4b, and G894T polymorphisms and male infertility: study for idiopathic asthenozoospermia and meta-analysis. Biol Reprod 2015;92:38.

31. Wu W, Lu J, Tang Q, Zhang S, Yuan B, Li J, et al. GSTM1 and GSTT1 null polymorphisms and male infertility risk: an updated meta-analysis encompassing 6934 subjects. Sci Rep 2013;3:2258.




32. Tang M, Wang S, Wang W, Cao Q, Qin C, Liu B, et al. The glutathione-S-transferase gene polymorphisms (GSTM1 and GSTT1) and idiopathic male infertility risk: a meta-analysis. Gene 2012;511:218–223.


33. Han LJ, He XF, Ye XH. Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and male infertility risk: an updated meta-analysis. Medicine (Baltimore) 2020;99:e23662.


34. Zou S, Li Z, Wang Y, Chen T, Song P, Chen J, et al. Association study between polymorphisms of PRMT6, PEX10, SOX5, and nonobstructive azoospermia in the Han Chinese population. Biol Reprod 2014;90:96.

35. van Rooij FJ, Qayyum R, Smith AV, Zhou Y, Trompet S, Tanaka T, et al. Genome-wide trans-ethnic meta-analysis identifies seven genetic loci influencing erythrocyte traits and a role for RBPMS in erythropoiesis. Am J Hum Genet 2017;100:51–63.



36. Terao C, Kawaguchi T, Dieude P, Varga J, Kuwana M, Hudson M, et al. Transethnic meta-analysis identifies GSDMA and PRDM1 as susceptibility genes to systemic sclerosis. Ann Rheum Dis 2017;76:1150–1158.



37. Tekola-Ayele F, Zhang C, Wu J, Grantz KL, Rahman ML, Shrestha D, et al. Trans-ethnic meta-analysis of genome-wide association studies identifies maternal ITPR1 as a novel locus influencing fetal growth during sensitive periods in pregnancy. PLoS Genet 2020;16:e1008747.



38. Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012;40:D930–D934.



39. Turner S, Armstrong LL, Bradford Y, Carlson CS, Crawford DC, Crenshaw AT, et al. Quality control procedures for genome-wide association studies. Curr Protoc Hum Genet 2011;Chapter 1:Unit1 19.




40. Higgins JP, Thomas J, Chandler J, Cumpston M, Li T, Page MJ, et al. Cochrane handbook for systematic reviews of interventions, version 6.3 (updated February 2022). London: Cochrane, 2022. Accessed 2022 Jul 26. Available from: http://www.training.cochrane.org/handbook.