|
 |
- Search
| Genomics Inform > Volume 19(4); 2021 > Article |
|
Abstract
In addition to mutations and copy number alterations, gene fusions are commonly identified in cancers. In thyroid cancer, fusions of important cancer-related genes have been commonly reported; however, extant panels do not cover all clinically important gene fusions. In this study, we aimed to develop a custom RNA-based sequencing panel to identify the key fusions in thyroid cancer. Our ThyChase panel was designed to detect 87 types of gene fusion. As quality control of RNA sequencing, five housekeeping genes were included in this panel. When we applied this panel for the analysis of fusions containing reference RNA (HD796), three expected fusions (EML4-ALK, CCDC6-RET, and TPM3-NTRK1) were successfully identified. We confirmed the fusion breakpoint sequences of the three fusions from HD796 by Sanger sequencing. Regarding the limit of detection, this panel could detect the target fusions from a tumor sample containing a 1% fusion-positive tumor cellular fraction. Taken together, our ThyChase panel would be useful to identify gene fusions in the clinical field.
Thyroid cancer is one of the most rapidly increasing cancers throughout the world, including South Korea [1]. Although most thyroid cancers show more favorable behavior than other cancers, and the 5-year disease-specific survival of thyroid cancer is above 98% [2], some thyroid cancers show aggressive behavior such as distant metastasis [3].
Next-generation sequencing (NGS) technology and efforts to identify genetic alterations in cancers, such as the Cancer Genome Atlas, have revealed genetic alteration profiles in diverse cancers [4]. In addition to mutations and copy number alterations, gene fusions are commonly identified in cancers, including thyroid cancer [5,6]. Gene fusions are mainly caused by chromosomal rearrangement; therefore, fusion events may have more tumorigenic implications than point mutations because cancer-related genes such as the RET oncogene can be overactivated through gene fusion [7]. The most common fusion events in thyroid cancer are the RET/PTC rearrangement in papillary thyroid cancer (10%–30%) and PPARG-PAX8 rearrangement in follicular thyroid cancer (30%–60%) [8]. Various other gene fusions have also been identified in thyroid cancer, including RET, THADA, NTRK1, NTRK3, ALK, BRAF, MET, and FGFR2 [8]. It is well known that gene fusions can affect the tumor behavior and prognosis of thyroid cancer [9,10]. For example, NTRK1/3 fusions have been reported to be associated with advanced tumor stage and aggressive lymphovascular invasion [11-13]. Tumors with ALK fusions have been suggested to have a higher likelihood of dedifferentiation [14]. Therefore, detecting gene fusion events is essential both for diagnostic purposes and for predicting patients’ prognoses.
From a technical standpoint, the detection of fusion genes by DNA-based NGS is almost impossible due to the presence of diverse-sized intronic sequences between the fusion target exons. Therefore, RNA-based NGS panel analyses are commonly conducted to detect the target fusions in thyroid cancer in addition to the use of DNA-based NGS to detect somatic mutations. RNA-based NGS panels should include housekeeping genes. Since housekeeping genes are expressed in all tissue compartments and cell types, they can be used for quality control and normalization of NGS data [15]. In addition, housekeeping genes may drive the expression of fusion genes such as VIT-ALK in lung adenocarcinoma [16,17]. Multiple panels for thyroid molecular analysis have been developed [9]. Among them, ThyroSeq, a DNA- and RNA-based NGS assay including 112 genes, is the most commonly used panel across the world; it can detect more than 100 genetic alterations, including major gene mutations, fusions, and gene expression alterations [18]. This panel provides high accuracy for detecting all common types of thyroid cancer and parathyroid lesions using a fine-needle aspiration sample. However, no extant panels cover all clinically important gene fusions in thyroid cancer.
In this study, we aimed to develop a custom RNA-based NGS panel to identify the important fusion events in thyroid cancer. In addition to the key fusions in thyroid cancer, uncommonly reported fusions and fusion subtypes were also included in this panel.
In this study, we used two standard materials: HD796 (Horizon Discovery, Cambridge, UK) as a fusion-positive control and HD783 (Horizon Discovery) as a fusion-negative control. HD796 is a formalin-fixed paraffin-embedded (FFPE) tissue that contains the EML4-ALK, CCDC6-RET, SLC34A2-ROS1, TPM3-NTRK1, and ETV6-NTRK3 fusions. HD783 is an FFPE sample that does not contain those fusions. RNA was extracted from the FFPE sample using an FFPE Total RNA Miniprep System kit (Promega, Madison, WI, USA). The quality and quantity of RNA samples were determined using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
cDNAs were synthesized using a SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific) and used for NGS library preparation. The libraries were manually constructed using our custom thyroid fusion panel, ThyChase. The amplicon library was prepared with the Ion Plus Fragment Library Kit (Life Technologies, Waltham, MA, USA) and the Ion Xpress Barcode Adapters Kit (Life Technologies) according to the manufacturer's instructions. In detail, 10 μL of cDNA was amplified in reaction mixtures of 59 μL containing 45 μL of Platinum PCR SuperMix High Fidelity and 4 μL of ThyChase panel. Polymerase chain reaction (PCR) was performed with a GeneAmp 9700 thermal cycler (Thermo Fisher Scientific) under the conditions of 95°C for 2 min followed by 35 cycles of 95°C for 15 s, 58°C for 15 s, 68°C for 10 s, and a final hold at 4°C. Libraries were purified using 106 μL of AMPure XP Reagent (Beckman Coulter, Miami, FL, USA) on a magnetic stand (Thermo Fisher Scientific) and eluted with 25 μL of low tris-EDTA buffer. Then, adapter ligation and nick repairing were performed to make barcode sequencing adapters (Ion Xpress Barcode Adapters, Thermo Fisher Scientific). Finally, the libraries were quantified using quantitative PCR (qPCR; Ion Library Quantitation Kit, Thermo Fisher Scientific) on a QuantStudio 12K Flex Real-Time PCR System qPCR machine (Thermo Fisher Scientific).
Emulsion PCR, bead enrichment, and chip loading procedures were automatically performed on an Ion Chef instrument (Thermo Fisher Scientific) using Ion 510, 520, and 530 Kits (Thermo Fisher Scientific). A planned run was created for each chip within Ion Torrent Suite Software v5.12.1 (Thermo Fisher Scientific) with the template size set at 200 bp. The NGS libraries were then sequenced on an Ion S5 XL sequencer (Thermo Fisher Scientific) [18].
Raw sequence data were analyzed with the Torrent Suite (version 5.12.1, Thermo Fisher Scientific). A custom reference genome was assembled to contain sequences of the 87 designed target fusions and five housekeeping genes based on hg19. To call the mapped sequence data, we used Torrent Coverage Analysis (version 5.12.0.0). More than five support reads were considered as fusion-positive. The identified fusions were then manually inspected in the Integrative Genomics Viewer (IGV, Broad Institute, Cambridge, MA, USA). The mean sequencing depth was 5,189× (range, 3,665× to 6,729×) across the entire target region (Supplementary Table 1). The dataset for the current study is available from the corresponding author upon reasonable request.
To determine the limit of detection (LOD) of fusions, we diluted the RNA extracted from the NCI-H2228 cell line (EML4-ALK fusion-positive) by mixing it with the RNA extracted from the FTC-133 cell line (EML4-ALK fusion-negative) from 100% to 0.5%. The RNAs were subjected to RNA sequencing using the ThyChase panel. To verify the fusions identified by the ThyChase panel, we performed Sanger sequencing of the fusion amplicons.
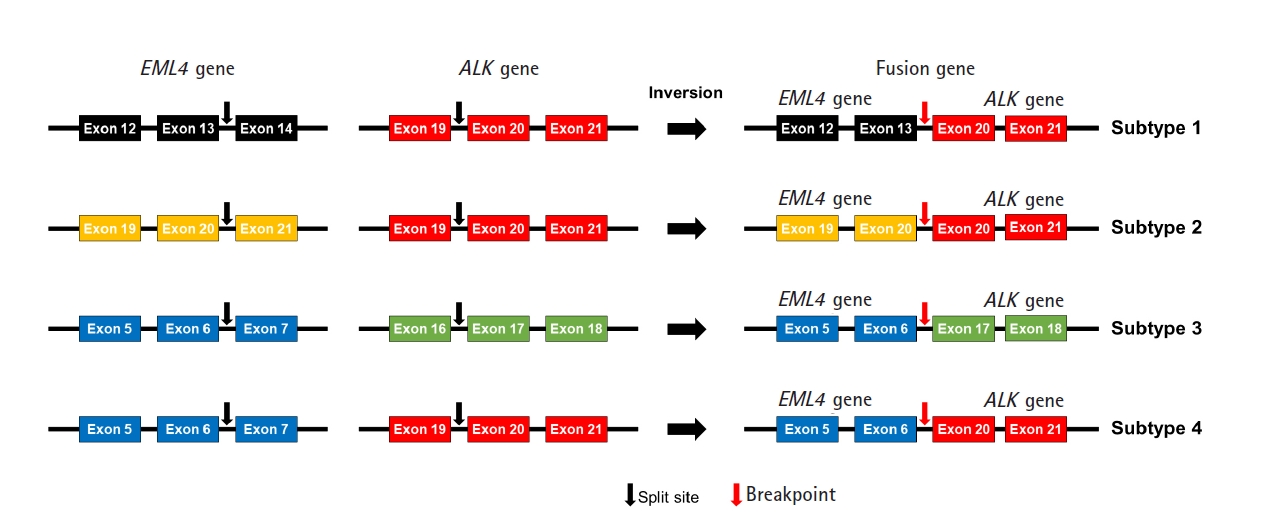
We designed an NGS panel named ThyChase containing 92 genes, targeting 87 gene fusion types and five housekeeping genes. The fusion targets were selected based on previous reports and the COSMIC database [7,8,19-64]. Of the 15 fusion targets, eight (RET, THADA, BRAF, ALK, FGFR2, NTRK1, NTRK3, and PPARG) are known to have multiple fusion partners, while the other seven are known to have a single fusion partner. Details of the fusion genes and their fusion partners are listed in Table 1. In some fusions, there are different fusion breakpoints, although the fusion partners are the same. For example, this panel can detect four fusion breakpoints of EML4-ALK fusion (exon 13 of EML4 - exon 20 of ALK, exon 20 of EML4 - exon 20 of ALK, exon 6 of EML4 - exon 17 of ALK, and exon 6 of EML4 - exon 20 of ALK) (Fig. 1). In total, 27 fusion subtypes can be discriminated with this panel (Supplementary Table 2). In addition to gene fusion, ThyChase includes five housekeeping genes for quality control of the experimental procedures of RNA sequencing and analysis: CHMP2A, JUN, FBXW2, MET, and PUM1.
We used HD796 RNA as a fusion-positive standard material. HD796 contains EML4-ALK, CCDC6-RET, TPM3-NTRK1, SLC34A2-ROS1, and ETV6-NTRK3 gene fusions [65]. Of the five fusions, three (EML4-ALK, CCDC6-RET, and TPM3-NTRK1) were included in our ThyChase panel. The ETV6-NTRK3 gene fusion was also included in ThyChase; however, the ETV6-NTRK3 fusion breakpoint of the HD796 RNA was different from the fusion breakpoint covered by ThyChase. Therefore, we targeted the three gene fusions for optimization and validation of ThyChase. HD783 RNA was used as a fusion-negative control that did not harbor any of the above-mentioned gene fusions [65].
Before detecting the gene fusions, as technical validation, we checked whether the RNA expression of the housekeeping genes included in this panel could be stably detected under various experimental conditions with the HD796 RNA. As expected, expression of the housekeeping genes was stably detected and the read counts mapped to each target gene were above 102, suggesting that our custom thyroid gene fusion panel is suitable for RNA sequencing (Fig. 2A). In parallel, to optimize the library preparation, we applied six different amplification conditions: two different amounts of template RNA were applied (10 ng and 100 ng) with three different primer concentrations (62.5, 125, and 187.5 nM). All five genes showed similar levels of the sequencing read counts in the six conditions (Fig. 2A). Therefore, we set the reaction condition as 10 ng of template RNA and 187.5 nM of primers. Regarding the quality control of ThaChase, we set >102 read counts for every housekeeping gene as a threshold of a reliable RNA sequencing reaction, meaning that we could interpret the gene fusions identified by this panel sequencing analysis as true when the read counts of all housekeeping genes were above 102.
We next examined the three (EML4-ALK, CCDC6-RET, and TPM3-NTRK1) fusions in the HD796 reference RNA. RNA panel sequencing was performed based on the optimized reaction conditions described above. As expected, all three fusion targets were successfully detected in the HD796 RNA, whereas they were not identified in the HD783 RNA (Fig. 2B). When we checked the read count of the housekeeping genes, all the target genes had >102 read counts (Supplementary Fig. 1), suggesting that the fusions detected by our thyroid gene fusion panel were reliable. These data also support the specificity of the fusions detected by our ThyChase panel.
Next, we applied ThyChase to cell lines. For this, we used the RNA extracted from a lung cancer cell line (NCI-H2228), which is known to harbor the EML4-ALK fusion [66]. As expected, the EML4-ALK fusion was successfully detected in the NCI-H2228 cell line (Fig. 3A). Through this experiment, we confirmed that the ThyChase panel could identify the target fusions from cancer cell lines in addition to the fusion-positive reference RNA. This result suggests that our system is applicable for cancer samples.
To determine the LOD of the ThyChase panel for calling fusions with high confidence, the lowest tumor percent with high-confidence detection was examined. To achieve this, we analyzed NCI-H2228 (EML4-ALK fusion-positive) samples that were diluted with FTC-133 (EML4-ALK fusion-negative) by different dilution factors (100% to 0.5%). As a result, the read count of EML4-ALK fusion decreased in a dose-dependent manner from 100% to 0.5%, and we could identify fusion-supporting reads from the 0.5% fusion-positive tumor cellular fraction (Fig. 3B). However, the lowest percentage satisfying our high-confidence fusion calling criterion (>5 fusion-supporting reads) was a >1% fusion-positive tumor cellular fraction. Therefore, a 1% tumor fraction was determined to be the LOD of our assay.
To verify the three fusions identified by our ThyChase panel from HD796 RNA, the RNA sequencing results were visualized using IGV. IGV showed the CCDC6-RET fusion breakpoint where exon 1 of CCDC6 was fused with exon 12 of RET (Fig. 4A). In the EML4-ALK fusion, exon 13 of EML4 was fused with exon 20 of ALK (Fig. 4B). In the breakpoint of TPM3-NTRK1 fusion, exon 7 of TPM3 and exon 10 of NTRK1 were fused (Fig. 4C). In addition, we confirmed the fusion subtype of EML4-ALK fusion, which was identified from NCI-H2228, where exon 6 of EML4 was fused with exon 20 of ALK (Fig. 4D).
As a final confirmation of the fusions and their breakpoints, we performed Sanger sequencing of the amplicons of the fusions from HD796. Sanger sequencing revealed the fusion breakpoint sequences of the EML4-ALK (exon 13 of EML4 and exon 20 of ALK), CCDC6-RET (exon 1 of CCDC6 and exon 12 of RET), and TPM3-NTRK1 (exon 7 of TPM3 and exon 10 of NTRK1) fusions (Fig. 5).
In conclusion, we developed an RNA-based sequencing panel focused on identifying fusions in thyroid cancer. The ThyChase panel was designed to detect 87 gene fusion types. As quality control for RNA sequencing, five housekeeping genes were included in this panel. When we applied this panel for the analysis of fusions contained in the reference RNA (HD796), the three expected fusions (EML4-ALK, CCDC6-RET, and TPM3-NTRK1) were successfully identified. We also confirmed that this fusion-focused panel could identify the target fusions from a cancer cell line in addition to the fusion-positive reference RNA. In terms of the LOD, this panel could detect the target fusions from a tumor sample containing a 1% fusion-positive tumor cellular fraction. We finally verified the fusion breakpoint sequences of the three fusions from HD796. Although we could not verify all of the designed fusions in this study due to limitations of the fusion reference materials, all the data in this study indicate that the ThyChase panel can reliably identify the key fusions in thyroid cancer. Taken together, the ThyChase panel would be useful to identify gene fusions in the clinical field.
Notes
Acknowledgments
This work was supported by a grant from the National Research Foundation of Korea (2017M3C9A6047615, 2019R1A5A2027588, 2019R1C1C1004909). We thank KREONET (Korea Research Environment Open NETwork) and KISTI (Korea Institute of Science and Technology Information) for allowing us to use their network infrastructure.
Supplementary Materials
Supplementary data can be found with this article online at http://www.genominfo.org.
Fig. 2.
Technical validation of RNA sequencing and detection of gene fusion. (A) RNA expression levels of the five housekeeping genes. We applied six different amplification conditions: two different amounts of template RNA were applied (10 ng and 100 ng) with three different primer concentrations (62.5, 125, and 187.5 nM). These six combinations are represented as different colors in the plot. The X-axis represents the gene name; the Y-axis represents read counts. (B) Identification of the fusions (EML4-ALK, CCDC6-RET, and TPM3-NTRK1) from the HD796 and HD783 RNAs. The Y-axis represents read counts.

Fig. 3.
Identification of EML4-ALK fusion from the NCI-H2228 cell line and limits of detection (LOD). (A) Identification of the EML4-ALK fusion. All five housekeeping genes showed >102 read counts. (B) To determine the LOD of the fusion, we diluted the NCI-H2228 RNA from 100% to 0.5% and performed RNA sequencing.
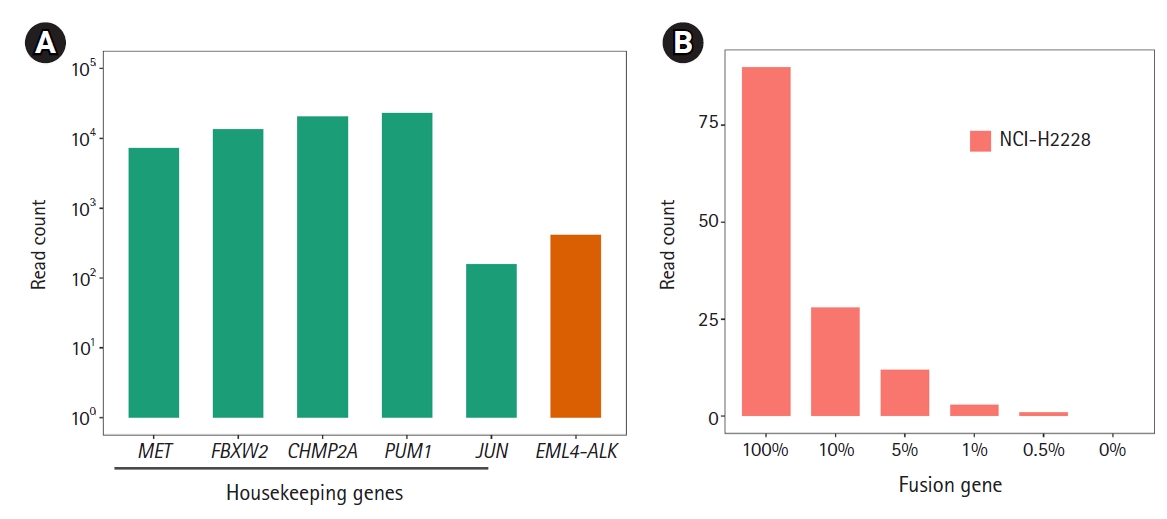
Fig. 4.
Integrative Genomics Viewer plot of the fusion breakpoints. (A) CCDC6-RET fusion breakpoint (exon 1 of CCDC6 and exon 12 of RET). (B) EML4-ALK fusion breakpoint (exon 13 of EML4 and exon 20 of ALK). (3) TPM3-NTRK1 fusion breakpoint (exon 7 of TPM3 and exon 10 of NTRK1). (4) EML4-ALK fusion breakpoint (exon 6 of EML4 and exon 20 of ALK).

Fig. 5.
Confirmation of the fusion breakpoints by Sanger sequencing. Vertical red lines represent fusion breakpoints.
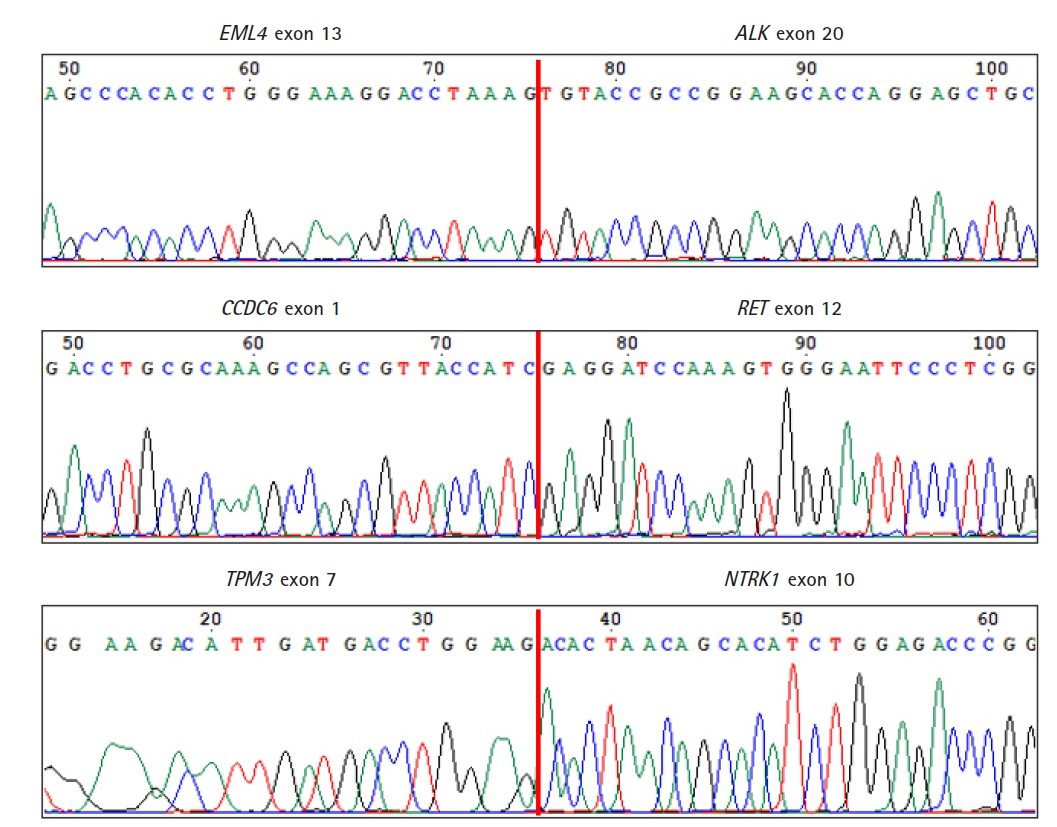
Table 1.
The fusion genes and their fusion partners contained in the panel
| Fusion gene | Partner gene(s) | Reference |
|---|---|---|
| THADA | IGF2BP3, LOC389473, LOC100505678, TRA2A | [5,19,46,47,54] |
| RET | CCDC6, ERC1, FKBP15, GOLGA5, HOOK3, KIAA1217, KTN1, NCOA4, PCM1, PRKAR1A, TRIM24, TRIM33, TRIM27, SPECC1L, TBL1XR1, AKAP13, DLG5, SQSTM1, CCDC186, AFAP1L2, PPFIBP2, KIF5B, | [7,20-39,44,46,54] |
| BRAF | AKAP9, AGK, LMO7, BCL2L11, CCNY, FAM114A2, OSBPL1A, OSBPL9, MACF1, POR, SND1, MKRN1, ZC3HAV1, PICALM, NFYA, AP3B1 | [23,40-47,54] |
| ALK | STRN, EML4, GFPT1, GTF2IRD1, CCDC149, | [23,48-53] |
| FGFR2 | WARS, KIAA1598, OFD1, VCL | [46,47,59] |
| NTRK1 | IRF2BP2, TFG, TPM3, TPR, SQSTM1, SSBP2 | [46,47,54-58] |
| NTRK3 | ETV6, RBPMS, SQSTM1, EML4 | [41,47,54,60] |
| PPARG | CREB3L2, PAX8 | [8,41,61,62] |
| UACA | LTK | [47] |
| MET | TFG | [23] |
| SS18 | SLC5A11 | [63] |
| RNF213 | SLC26A11 | [46] |
| ROS1 | CCDC30 | [64] |
| RAF1 | AGGF1 | [23] |
| EZR | ERBB4 | [46] |
References
1. Kilfoy BA, Zheng T, Holford TR, Han X, Ward MH, Sjodin A, et al. International patterns and trends in thyroid cancer incidence, 1973-2002. Cancer Causes Control 2009;20:525–531.


2. Wang TS, Sosa JA. Thyroid surgery for differentiated thyroid cancer: recent advances and future directions. Nat Rev Endocrinol 2018;14:670–683.


3. Sampson E, Brierley JD, Le LW, Rotstein L, Tsang RW. Clinical management and outcome of papillary and follicular (differentiated) thyroid cancer presenting with distant metastasis at diagnosis. Cancer 2007;110:1451–1456.


4. Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018;173:371–385.


5. Gao Q, Liang WW, Foltz SM, Mutharasu G, Jayasinghe RG, Cao S, et al. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep 2018;23:227–238.


6. Demircioglu D, Cukuroglu E, Kindermans M, Nandi T, Calabrese C, Fonseca NA, et al. A Pan-cancer transcriptome analysis reveals pervasive regulation through alternative promoters. Cell 2019;178:1465–1477.


7. Ciampi R, Giordano TJ, Wikenheiser-Brokamp K, Koenig RJ, Nikiforov YE. HOOK3-RET: a novel type of RET/PTC rearrangement in papillary thyroid carcinoma. Endocr Relat Cancer 2007;14:445–452.


8. Nikiforova MN, Biddinger PW, Caudill CM, Kroll TG, Nikiforov YE. PAX8-PPARgamma rearrangement in thyroid tumors: RT-PCR and immunohistochemical analyses. Am J Surg Pathol 2002;26:1016–1023.

9. Nikiforov YE. Role of molecular markers in thyroid nodule management: Then and Now. Endocr Pract 2017;23:979–988.


10. Liu M, Chen P, Hu HY, Ou-Yang DJ, Khushbu RA, Tan HL, et al. Kinase gene fusions: roles and therapeutic value in progressive and refractory papillary thyroid cancer. J Cancer Res Clin Oncol 2021;147:323–337.


11. Prasad ML, Vyas M, Horne MJ, Virk RK, Morotti R, Liu Z, et al. NTRK fusion oncogenes in pediatric papillary thyroid carcinoma in northeast United States. Cancer 2016;122:1097–1107.


12. Chu YH, Dias-Santagata D, Farahani AA, Boyraz B, Faquin WC, Nose V, et al. Clinicopathologic and molecular characterization of NTRK-rearranged thyroid carcinoma (NRTC). Mod Pathol 2020;33:2186–2197.



13. Albert CM, Davis JL, Federman N, Casanova M, Laetsch TW. TRK fusion cancers in children: a clinical review and recommendations for screening. J Clin Oncol 2019;37:513–524.


14. Panebianco F, Nikitski AV, Nikiforova MN, Kaya C, Yip L, Condello V, et al. Characterization of thyroid cancer driven by known and novel ALK fusions. Endocr Relat Cancer 2019;26:803–814.



15. Wang Z, Lyu Z, Pan L, Zeng G, Randhawa P. Defining housekeeping genes suitable for RNA-seq analysis of the human allograft kidney biopsy tissue. BMC Med Genomics 2019;12:86.



16. Ni Chin WH, Li Z, Jiang N, Lim EH, Suang Lim JY, Lu Y, et al. Practical considerations for using RNA sequencing in management of B-lymphoblastic leukemia: Malaysia-Singapore Acute Lymphoblastic Leukemia 2020 Implementation Strategy. J Mol Diagn 2021;23:1359–1372.


17. Hu S, Li Q, Peng W, Feng C, Zhang S, Li C. VIT-ALK, a novel alectinib-sensitive fusion gene in lung adenocarcinoma. J Thorac Oncol 2018;13:e72–e74.


18. Wang L, Chen M, Wu B, Liu YC, Zhang GF, Jiang L, et al. Massively parallel sequencing of forensic STRs using the Ion Chef and the Ion S5 XL systems. J Forensic Sci 2018;63:1692–1703.


19. Panebianco F, Kelly LM, Liu P, Zhong S, Dacic S, Wang X, et al. THADA fusion is a mechanism of IGF2BP3 activation and IGF1R signaling in thyroid cancer. Proc Natl Acad Sci U S A 2017;114:2307–2312.



20. Di Cristofaro J, Marcy M, Vasko V, Sebag F, Fakhry N, Wynford-Thomas D, et al. Molecular genetic study comparing follicular variant versus classic papillary thyroid carcinomas: association of N-ras mutation in codon 61 with follicular variant. Hum Pathol 2006;37:824–830.


21. Santarpia L, Myers JN, Sherman SI, Trimarchi F, Clayman GL, El-Naggar AK. Genetic alterations in the RAS/RAF/mitogen-activated protein kinase and phosphatidylinositol 3-kinase/Akt signaling pathways in the follicular variant of papillary thyroid carcinoma. Cancer 2010;116:2974–2983.


22. Sheils OM, O'Leary JJ, Sweeney EC. Assessment of ret/PTC-1 rearrangements in neoplastic thyroid tissue using TaqMan RT-PCR. J Pathol 2000;192:32–36.


23. Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun 2014;5:4846.


24. Giannini R, Salvatore G, Monaco C, Sferratore F, Pollina L, Pacini F, et al. Identification of a novel subtype of H4-RET rearrangement in a thyroid papillary carcinoma and lymph node metastasis. Int J Oncol 2000;16:485–489.


25. Musholt PB, Imkamp F, von Wasielewski R, Schmid KW, Musholt TJ. RET rearrangements in archival oxyphilic thyroid tumors: new insights in tumorigenesis and classification of Hurthle cell carcinomas? Surgery 2003;134:881–889.


26. Elisei R, Romei C, Soldatenko PP, Cosci B, Vorontsova T, Vivaldi A, et al. New breakpoints in both the H4 and RET genes create a variant of PTC-1 in a post-Chernobyl papillary thyroid carcinoma. Clin Endocrinol (Oxf) 2000;53:131–136.


27. Liu RT, Chou FF, Wang CH, Lin CL, Chao FP, Chung JC, et al. Low prevalence of RET rearrangements (RET/PTC1, RET/PTC2, RET/PTC3, and ELKS-RET) in sporadic papillary thyroid carcinomas in Taiwan Chinese. Thyroid 2005;15:326–335.


28. Nakata T, Kitamura Y, Shimizu K, Tanaka S, Fujimori M, Yokoyama S, et al. Fusion of a novel gene, ELKS, to RET due to translocation t(10;12)(q11;p13) in a papillary thyroid carcinoma. Genes Chromosomes Cancer 1999;25:97–103.


29. Klugbauer S, Demidchik EP, Lengfelder E, Rabes HM. Detection of a novel type of RET rearrangement (PTC5) in thyroid carcinomas after Chernobyl and analysis of the involved RET-fused gene RFG5. Cancer Res 1998;58:198–203.

30. Hamatani K, Eguchi H, Ito R, Mukai M, Takahashi K, Taga M, et al. RET/PTC rearrangements preferentially occurred in papillary thyroid cancer among atomic bomb survivors exposed to high radiation dose. Cancer Res 2008;68:7176–7182.


31. Sheu SY, Schwertheim S, Worm K, Grabellus F, Schmid KW. Diffuse sclerosing variant of papillary thyroid carcinoma: lack of BRAF mutation but occurrence of RET/PTC rearrangements. Mod Pathol 2007;20:779–787.


32. Liu S, Gao A, Zhang B, Zhang Z, Zhao Y, Chen P, et al. Assessment of molecular testing in fine-needle aspiration biopsy samples: an experience in a Chinese population. Exp Mol Pathol 2014;97:292–297.


33. Corvi R, Berger N, Balczon R, Romeo G. RET/PCM-1: a novel fusion gene in papillary thyroid carcinoma. Oncogene 2000;19:4236–4242.


34. Cheung CC, Boerner SL, MacMillan CM, Ramyar L, Asa SL. Hyalinizing trabecular tumor of the thyroid: a variant of papillary carcinoma proved by molecular genetics. Am J Surg Pathol 2000;24:1622–1626.


35. Chua EL, Wu WM, Tran KT, McCarthy SW, Lauer CS, Dubourdieu D, et al. Prevalence and distribution of ret/ptc 1, 2, and 3 in papillary thyroid carcinoma in New Caledonia and Australia. J Clin Endocrinol Metab 2000;85:2733–2739.


36. Klugbauer S, Rabes HM. The transcription coactivator HTIF1 and a related protein are fused to the RET receptor tyrosine kinase in childhood papillary thyroid carcinomas. Oncogene 1999;18:4388–4393.


37. Saenko V, Rogounovitch T, Shimizu-Yoshida Y, Abrosimov A, Lushnikov E, Roumiantsev P, et al. Novel tumorigenic rearrangement, Delta rfp/ret, in a papillary thyroid carcinoma from externally irradiated patient. Mutat Res 2003;527:81–90.


38. Iyama K, Matsuse M, Mitsutake N, Rogounovitch T, Saenko V, Suzuki K, et al. Identification of three novel fusion oncogenes, SQSTM1/NTRK3, AFAP1L2/RET, and PPFIBP2/RET, in thyroid cancers of young patients in Fukushima. Thyroid 2017;27:811–818.


39. Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012;18:375–377.



40. Ciampi R, Knauf JA, Kerler R, Gandhi M, Zhu Z, Nikiforova MN, et al. Oncogenic AKAP9-BRAF fusion is a novel mechanism of MAPK pathway activation in thyroid cancer. J Clin Invest 2005;115:94–101.



41. Ricarte-Filho JC, Li S, Garcia-Rendueles ME, Montero-Conde C, Voza F, Knauf JA, et al. Identification of kinase fusion oncogenes in post-Chernobyl radiation-induced thyroid cancers. J Clin Invest 2013;123:4935–4944.



42. Cordioli MI, Moraes L, Carvalheira G, Sisdelli L, Alves MT, Delcelo R, et al. AGK-BRAF gene fusion is a recurrent event in sporadic pediatric thyroid carcinoma. Cancer Med 2016;5:1535–1541.



43. He H, Li W, Yan P, Bundschuh R, Killian JA, Labanowska J, et al. Identification of a recurrent LMO7-BRAF fusion in papillary thyroid carcinoma. Thyroid 2018;28:748–754.



44. Ibrahimpasic T, Xu B, Landa I, Dogan S, Middha S, Seshan V, et al. Genomic alterations in fatal forms of non-anaplastic thyroid cancer: identification of MED12 and RBM10 as novel thyroid cancer genes associated with tumor virulence. Clin Cancer Res 2017;23:5970–5980.



45. Efanov AA, Brenner AV, Bogdanova TI, Kelly LM, Liu P, Little MP, et al. Investigation of the relationship between radiation dose and gene mutations and fusions in post-chernobyl thyroid cancer. J Natl Cancer Inst 2018;110:371–378.


46. Yoo SK, Lee S, Kim SJ, Jee HG, Kim BA, Cho H, et al. Comprehensive analysis of the transcriptional and mutational landscape of follicular and papillary thyroid cancers. PLoS Genet 2016;12:e1006239.



47. Hu X, Wang Q, Tang M, Barthel F, Amin S, Yoshihara K, et al. TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res 2018;46:D1144–D1149.


48. Perot G, Soubeyran I, Ribeiro A, Bonhomme B, Savagner F, Boutet-Bouzamondo N, et al. Identification of a recurrent STRN/ALK fusion in thyroid carcinomas. PLoS One 2014;9:e87170.



49. Kelly LM, Barila G, Liu P, Evdokimova VN, Trivedi S, Panebianco F, et al. Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc Natl Acad Sci U S A 2014;111:4233–4238.



50. Hamatani K, Mukai M, Takahashi K, Hayashi Y, Nakachi K, Kusunoki Y. Rearranged anaplastic lymphoma kinase (ALK) gene in adult-onset papillary thyroid cancer amongst atomic bomb survivors. Thyroid 2012;22:1153–1159.



51. Zeng Q, Gao H, Zhang L, Qin S, Gu Y, Chen Q. Coexistence of a secondary STRN-ALK, EML4-ALK double-fusion variant in a lung adenocarcinoma patient with EGFR mutation: a case report. Anticancer Drugs 2021;32:890–893.


52. Ji JH, Oh YL, Hong M, Yun JW, Lee HW, Kim D, et al. Identification of driving ALK fusion genes and genomic landscape of medullary thyroid cancer. PLoS Genet 2015;11:e1005467.



53. Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest 2016;126:1052–1066.



54. Liang J, Cai W, Feng D, Teng H, Mao F, Jiang Y, et al. Genetic landscape of papillary thyroid carcinoma in the Chinese population. J Pathol 2018;244:215–226.


55. Greco A, Mariani C, Miranda C, Lupas A, Pagliardini S, Pomati M, et al. The DNA rearrangement that generates the TRK-T3 oncogene involves a novel gene on chromosome 3 whose product has a potential coiled-coil domain. Mol Cell Biol 1995;15:6118–6127.



56. Butti MG, Bongarzone I, Ferraresi G, Mondellini P, Borrello MG, Pierotti MA. A sequence analysis of the genomic regions involved in the rearrangements between TPM3 and NTRK1 genes producing TRK oncogenes in papillary thyroid carcinomas. Genomics 1995;28:15–24.


57. Greco A, Miranda C, Pagliardini S, Fusetti L, Bongarzone I, Pierotti MA. Chromosome 1 rearrangements involving the genes TPR and NTRK1 produce structurally different thyroid-specific TRK oncogenes. Genes Chromosomes Cancer 1997;19:112–123.


58. Greco A, Pierotti MA, Bongarzone I, Pagliardini S, Lanzi C, Della Porta G. TRK-T1 is a novel oncogene formed by the fusion of TPR and TRK genes in human papillary thyroid carcinomas. Oncogene 1992;7:237–242.

59. Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013;3:636–647.



60. Leeman-Neill RJ, Kelly LM, Liu P, Brenner AV, Little MP, Bogdanova TI, et al. ETV6-NTRK3 is a common chromosomal rearrangement in radiation-associated thyroid cancer. Cancer 2014;120:799–807.


61. Lui WO, Zeng L, Rehrmann V, Deshpande S, Tretiakova M, Kaplan EL, et al. CREB3L2-PPARgamma fusion mutation identifies a thyroid signaling pathway regulated by intramembrane proteolysis. Cancer Res 2008;68:7156–7164.


62. Chia WK, Sharifah NA, Reena RM, Zubaidah Z, Clarence-Ko CH, Rohaizak M, et al. Fluorescence in situ hybridization analysis using PAX8- and PPARG-specific probes reveals the presence of PAX8-PPARG translocation and 3p25 aneusomy in follicular thyroid neoplasms. Cancer Genet Cytogenet 2010;196:7–13.


63. Kasaian K, Wiseman SM, Walker BA, Schein JE, Zhao Y, Hirst M, et al. The genomic and transcriptomic landscape of anaplastic thyroid cancer: implications for therapy. BMC Cancer 2015;15:984.



64. Ritterhouse LL, Wirth LJ, Randolph GW, Sadow PM, Ross DS, Liddy W, et al. ROS1 rearrangement in thyroid cancer. Thyroid 2016;26:794–797.


65. Nohr E, Kunder CA, Jones C, Sutton S, Fung E, Zhu H, et al. Development and clinical validation of a targeted RNAseq panel (Fusion-STAMP) for diagnostic and predictive gene fusion detection in solid tumors. Preprint at https://www.biorxiv.org/content/10.1101/870634v1.full (2019).

- TOOLS
- Related articles in GNI