Introduction
From the beginning of endosymbiosis between the origin eukaryote cell and alphaproteobacteria, the phenomenon of mitochondrial gene transfer to the host cell is still an ongoing evolutionary process [1,2]. It is termed nuclear mitochondrial DNA (Numt), pronounced "new might" [3]. In general, the mutation rate of nuclear DNA is lower than that of the mitochondrial genome. For this reason, Numt is often called a molecular fossil and is used as a molecular marker for speciation events in evolution [4,5]. While Numt insertion is a well-known phenomenon, the mechanism of DNA insertion into the nuclear genome is not clear. One of the strongly supported hypotheses is that during the process of double-strand break repair, an absorbed mitochondrial DNA fragment is inserted into the nuclear genome via a non-homologous end joining event [6,7,8,9].
After whole-genome sequencing was finished, Numt analysis was performed in various species: cat, cattle, dog, fruit fly, gorilla, grasshopper, goose, horse, horseshoe bat, honeybee, human, maize, squirrel, and whale [10,11,12,13,14,15,16,17]. One review paper summarized all existences of Numt in complete genome sequences [18]. In the case of whale, a phylogenetic analysis of Numts with six whale species was carried out, establishing Numt as an evolutionary marker in speciation events [19]. In plant, a recent study discovered that Numt insertion is dispersed throughout the periphery of the centromere [20]. But, there are many barriers in plant Numt analyses. Genomic complexity is a big problem in not only the nuclear genome but also the mitochondrial genome [21,22,23].
In this article, Numts of two green algae (Chlamydomonas reinhardtii and Coccomyxa subellipsoidea), three monocots (Oryza sativa, Sorghum bicolor, and Zea mays), and four eudicots (Vitis vinifera, Glycine max, Brassica rapa, and Arabidopsis thaliana), for which whole-genome nuclear and mitochondrial sequences are publically available, were detected using the nucleotide-nucleotide Basic Local Alignment Search Tool (BLASTN) searches and subjected to a basic analysis for their fundamental properties, which will be required in further comparative genome analyses in plants.
Methods
Data sources
We downloaded all of the genomic data and gene annotation data (gff3) of two green algae genomes (C. reinhardtii and C. subellipsoidea) from the Joint Genome Institute, four plant genomes (O. sativa, Z. mays, V. vinifera, and A. thaliana) from the Ensembl genome database, two plant genomes (G. max and S. bicolor) from the Plant Genome Database, and the B. rapa genome and annotation information data, available online from the B. rapa Database. We also collected each mitochondrial genome sequence from the National Center for Biotechnology Information (NCBI). All these data sources are summarized in Table 1.
Detection of Numts and data generation
Plant Numt insertions were identified using BLASTN local alignment tools in the BLAST program package (ver. 2.2.26), with mitochondrial genomic DNA as a query sequence and each genome dataset as a BLAST database. The execution options included an e-value cutoff set to 0.01, filtering switched off (-dust no), a mismatch penalty of ŌĆō2, and a word size of 9. The neighboring Numt hits within 10 kb were, if necessary, merged into a single event of Numt insertions. All of these analytical processes were carried out with in-house Python codes.
Calculation of odd ratio for Numt insertion loci
To calculate the relative abundance of each genomic feature, we gathered the length information of the categories, such as gene, coding sequence (CDS), exon, pseudogene, and noncoding RNA (ncRNA) (tRNA, rRNA, and long non-coding RNA [lncRNA]), from the gene annotation files of each species (gff3 format). The total length of exons and introns included only the protein-coding genes. The total length of introns was computed by subtracting the total exon length from the sum of all gene lengths. With all of the length information, we estimated the portion of each feature by dividing the total length of each feature (Si) by each whole-genome length (G). The relative abundance (RAi) of each feature was then calculated as follows: , where Ci is the count of the genic feature i in a species.
Results and Discussion
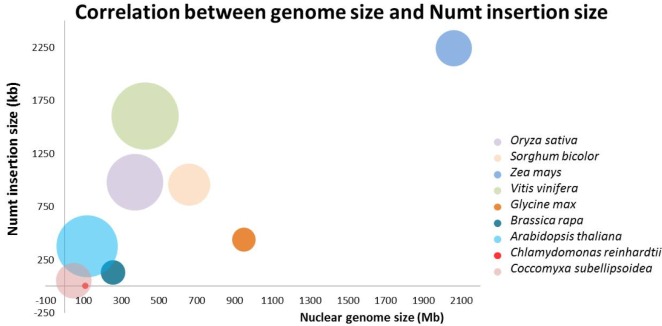
All of the data are summarized in Table 2. The genome size of the nine plants species varied from 48 Mb to 2 Gb. The mitochondria genome size also varied from 15.8 kb to 773 kb. There was no correlation between whole-genome size and mitochondrial whole-genome size. We drew a correlation chart between whole nuclear genome length and the sum of the inserted Numt lengths (Fig. 1). The larger the genome size, the more nuclear mitochondrial insertions there were. This confirms a previous study result [18]. The added green algae species also showed this tendency. One of the peculiarities of plant species is their many Numt hits. Except for green algae (C. reinhardtii and C. subellipsoidea), the number of BLAST hits after merging all overlapping hits ranged from 770 for A. thaliana to 14,509 for V. vinifera. Furthermore, when integrating all of the neighboring hits within 10 kb into one single event, the hit count ranged from 562 in A. thaliana to 9,022 in V. vinifera. This implies that the transposition of mitochondrial DNA of plants into chromosomal DNA is more preferable than in whale species [19].
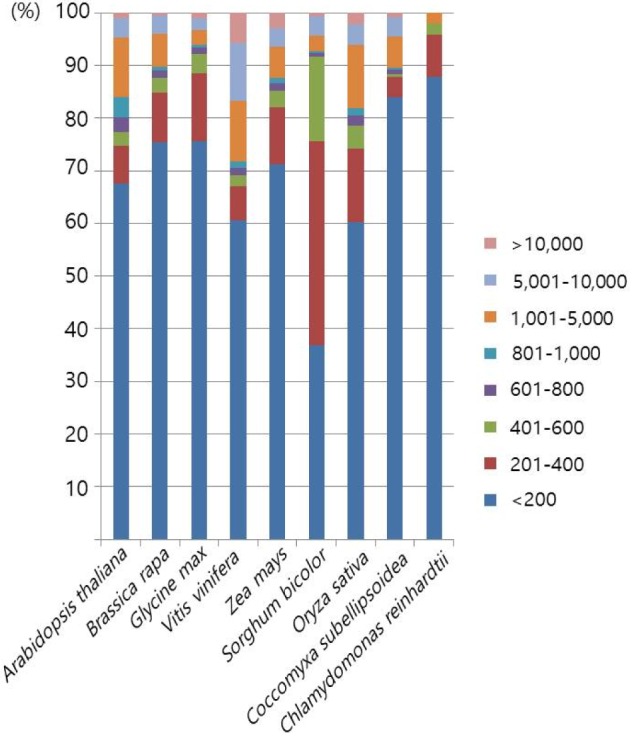
Next, we examined the size distribution of the inserted Numts. Here, we merged the neighboring hits within 10 kb into single events. The merged hits showed a high degree of variation in sizeŌĆöthe shortest and largest being 25 bp and 107 kb, respectively (Table 2). The size distribution of Numt was also quite variable between species (Fig. 2). Over 70% of Numts were less than 400 bp in all of the analyzed plants. Green algae species that had shorter mitochondrial DNA than other species had over 80% in the group with less than 200 bp, especially in C. reinhardtii (over 96%). V. vinifera, which has a larger mitochondrial genome size than other plants, included 30% of Numts over 1 kb in size, and half of this group was over 5 kb. Z. mays, which has the largest genome and the second largest mitochondrial genome, and B. rapa, which has a relatively shorter genome than Z. mays, showed similar ratio distributions. In general, species having short mitochondrial genomes had a large ratio of short Numts. When comparing monocots and eudicots, there was no clearly shared feature. But, there were some differences when contrasting green algae and land plants. However, it is not a matter of the species group but rather a matter of genomic size variation. There are two kinds of closely related speciation events: one is between A. thaliana and B. rapa, and the other is between S. bicolor and Z. mays. In each of the speciation events, there were whole-genome triplication or duplication events, leading to B. rapa and Z. mays [24,25]. Because of that, each pair has a similar genomic content, but the within-pair Numt size distribution patterns are different. In general, B. rapa and Z. mays have lower ratios of long sizes of Numts than A. thaliana and S. bicolor, respectively. Genome triplication or duplication events may have split the long Numt sequences, such that the number of long Numts was reduced. These patterns were also observed in the speciation between G. max and V. vinifera.
In the previous whale Numt study [19], they also performed a Numt size distribution analysis. The average whale genome size is 2.5 Gb, and the average mitochondrial genome size is 16 kb. It has a much larger nuclear genome and smaller mitochondrial genome. In whales, the Numt size group over 5 kb is under 2%, but in plants, it is over 4%, and in V. vinifera, it is over 17%. It is presumed that as a result of a 20-fold larger mitochondrial genome, even if going through the second whole-genome duplication event, there are longer Numt sequences that still reside in the plant genome.
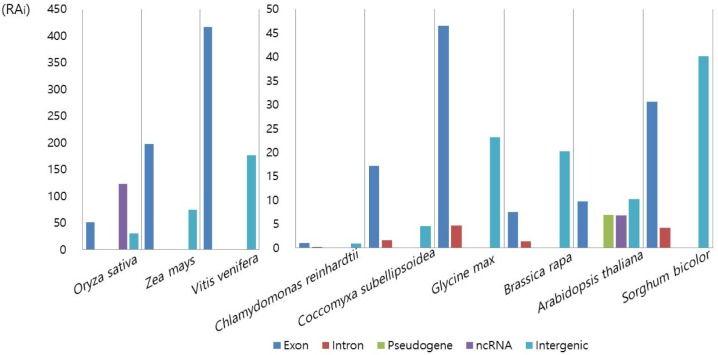
The next analysis was the classification of Numt insertion loci by genic features (Table 3). In land plants, a substantial portion of Numt hits lay in intergenic regions, except for green algae, where over 70% of the hits were found within genic boundaries. Within genic regions, over 90% of the hits overlapped exons. This is in contrast with the Numt hits in animals, like whales, where the total number of Numt hits was quite low and in which fewer hits were found in exons than in introns [19]. When we calculated the relative abundance of each genic feature after accounting for the total size of each genic feature, the exon was the most enriched in most plants (Fig. 3). Considering the importance of exons in biological processes, it may be tempting to speculate that the numerous Numt insertions into exons may affect the diversity of plant phenotypes.
Many research studies on Numt analysis have been performed. But, they usually lack details on Numts, such as the correlation between genome size and inserted Numt size, Numt size distribution ratio, loci classification by gene annotation, and so on. Our general basic analysis shows an interesting tendency but is still not enough to infer the biological meaning. Currently, not many plant genomes have been completely sequenced, and furthermore, their accuracy is somewhat compromised due to high repeat contents or high heterozygosity in the genomes. In order to draw a clearer picture of the effect of Numt insertion in the nuclear genome, more population-level genomic data and more accurate genome sequences may be required. Nevertheless, Numts may be one of the key clues of the mysterious biological implications of genomic analysis.